A nascent polypeptide domain that can regulate translation elongation
- PMID: 15020769
- PMCID: PMC384695
- DOI: 10.1073/pnas.0400554101
A nascent polypeptide domain that can regulate translation elongation
Abstract
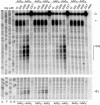
The evolutionarily conserved fungal arginine attenuator peptide (AAP), as a nascent peptide, stalls the translating ribosome in response to the presence of a high concentration of the amino acid arginine. Here we examine whether the AAP maintains regulatory function in fungal, plant, and animal cell-free translation systems when placed as a domain near the N terminus or internally within a large polypeptide. Pulse-chase analyses of the radiolabeled polypeptides synthesized in these systems indicated that wild-type AAP functions at either position to stall polypeptide synthesis in response to arginine. Toeprint analyses performed to map the positions of stalled ribosomes on transcripts introduced into the fungal system revealed that ribosome stalling required translation of the AAP coding sequence. The positions of the stalled ribosomes were consistent with the sizes of the radiolabeled polypeptide intermediates. These findings demonstrate that an internal polypeptide domain in a nascent chain can regulate eukaryotic translational elongation in response to a small molecule. Apparently the peptide-sensing features are conserved in fungal, plant, and animal ribosomes. These data provide precedents for translational strategies that would allow domains within nascent polypeptide chains to modulate gene expression.
Figures

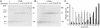

Similar articles
-
Sequence requirements for ribosome stalling by the arginine attenuator peptide.J Biol Chem. 2010 Dec 24;285(52):40933-42. doi: 10.1074/jbc.M110.164152. Epub 2010 Sep 30. J Biol Chem. 2010. PMID: 20884617 Free PMC article.
-
The arginine attenuator peptide interferes with the ribosome peptidyl transferase center.Mol Cell Biol. 2012 Jul;32(13):2396-406. doi: 10.1128/MCB.00136-12. Epub 2012 Apr 16. Mol Cell Biol. 2012. PMID: 22508989 Free PMC article.
-
The evolutionarily conserved eukaryotic arginine attenuator peptide regulates the movement of ribosomes that have translated it.Mol Cell Biol. 1998 Dec;18(12):7528-36. doi: 10.1128/MCB.18.12.7528. Mol Cell Biol. 1998. PMID: 9819438 Free PMC article.
-
Translation regulation via nascent polypeptide-mediated ribosome stalling.Curr Opin Struct Biol. 2016 Apr;37:123-33. doi: 10.1016/j.sbi.2016.01.008. Epub 2016 Feb 7. Curr Opin Struct Biol. 2016. PMID: 26859868 Review.
-
Nascent polypeptide sequences that influence ribosome function.Curr Opin Microbiol. 2011 Apr;14(2):160-6. doi: 10.1016/j.mib.2011.01.011. Epub 2011 Feb 20. Curr Opin Microbiol. 2011. PMID: 21342782 Review.
Cited by
-
Identification of novel Arabidopsis thaliana upstream open reading frames that control expression of the main coding sequences in a peptide sequence-dependent manner.Nucleic Acids Res. 2015 Feb 18;43(3):1562-76. doi: 10.1093/nar/gkv018. Epub 2015 Jan 23. Nucleic Acids Res. 2015. PMID: 25618853 Free PMC article.
-
Site-specific release of nascent chains from ribosomes at a sense codon.Mol Cell Biol. 2008 Jul;28(13):4227-39. doi: 10.1128/MCB.00421-08. Epub 2008 May 5. Mol Cell Biol. 2008. PMID: 18458056 Free PMC article.
-
Translational autoregulation of the S. cerevisiae high-affinity polyamine transporter Hol1.Mol Cell. 2021 Oct 7;81(19):3904-3918.e6. doi: 10.1016/j.molcel.2021.07.020. Epub 2021 Aug 9. Mol Cell. 2021. PMID: 34375581 Free PMC article.
-
Control of translation by eukaryotic mRNA transcript leaders-Insights from high-throughput assays and computational modeling.Wiley Interdiscip Rev RNA. 2021 May;12(3):e1623. doi: 10.1002/wrna.1623. Epub 2020 Aug 31. Wiley Interdiscip Rev RNA. 2021. PMID: 32869519 Free PMC article. Review.
-
A Conserved uORF Regulates APOBEC3G Translation and Is Targeted by HIV-1 Vif Protein to Repress the Antiviral Factor.Biomedicines. 2021 Dec 22;10(1):13. doi: 10.3390/biomedicines10010013. Biomedicines. 2021. PMID: 35052693 Free PMC article.
References
-
- Keenan, R. J., Freymann, D. M., Stroud, R. M. & Walter, P. (2001) Annu. Rev. Biochem. 70, 755–775. - PubMed
-
- Tenson, T. & Ehrenberg, M. (2002) Cell 108, 591–594. - PubMed
-
- Geballe, A. P. & Sachs, M. S. (2000) in Translational Control of Gene Expression, eds. Sonenberg, N., Hershey, J. W. B. & Mathews, M. B. (Cold Spring Harbor Lab. Press, Plainview, NY), pp. 595–614.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

