Characterization of severe acute respiratory syndrome-associated coronavirus (SARS-CoV) spike glycoprotein-mediated viral entry
- PMID: 15010527
- PMCID: PMC384725
- DOI: 10.1073/pnas.0306446101
Characterization of severe acute respiratory syndrome-associated coronavirus (SARS-CoV) spike glycoprotein-mediated viral entry
Abstract
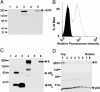
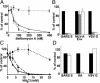
Severe acute respiratory syndrome-associated coronavirus (SARS-CoV) is a rapidly emerging pathogen with potentially serious consequences for public health. Here we describe conditions that result not only in the efficient expression of the SARS-CoV spike (S) protein on the surface of cells, but in its incorporation into lentiviral particles that can be used to transduce cells in an S glycoprotein-dependent manner. We found that although some primate cell lines, including Vero E6, 293T and Huh-7 cells, could be efficiently transduced by SARS-CoV S glycoprotein pseudoviruses, other cells lines were either resistant or very poorly permissive to virus entry. Infection by pseudovirions could be inhibited by several lysosomotropic agents, suggesting a requirement for acidification of endosomes for efficient S-mediated viral entry. In addition, we were able to develop a cell-cell fusion assay that could be used to monitor S glycoprotein-dependent membrane fusion. Although proteolysis did not enhance the infectivity of cell-free pseudovirions, trypsin activation is required for cell-cell fusion. Additionally, there was no apparent pH requirement for S glycoprotein-mediated cell-cell fusion. Together, these studies describe important tools that can be used to study SARS-CoV S glycoprotein structure and function, including approaches that can be used to identify inhibitors of the entry of SARS-CoV into target cells.
Figures
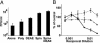
Similar articles
-
SARS coronavirus entry into host cells through a novel clathrin- and caveolae-independent endocytic pathway.Cell Res. 2008 Feb;18(2):290-301. doi: 10.1038/cr.2008.15. Cell Res. 2008. PMID: 18227861 Free PMC article.
-
A convenient cell fusion assay for the study of SARS-CoV entry and inhibition.IUBMB Life. 2006 Aug;58(8):480-6. doi: 10.1080/15216540600820974. IUBMB Life. 2006. PMID: 16916786 Free PMC article.
-
A single asparagine-linked glycosylation site of the severe acute respiratory syndrome coronavirus spike glycoprotein facilitates inhibition by mannose-binding lectin through multiple mechanisms.J Virol. 2010 Sep;84(17):8753-64. doi: 10.1128/JVI.00554-10. Epub 2010 Jun 23. J Virol. 2010. PMID: 20573835 Free PMC article.
-
The SARS-CoV S glycoprotein.Cell Mol Life Sci. 2004 Oct;61(19-20):2428-30. doi: 10.1007/s00018-004-4257-y. Cell Mol Life Sci. 2004. PMID: 15526150 Free PMC article. Review.
-
Cellular entry of the SARS coronavirus.Trends Microbiol. 2004 Oct;12(10):466-72. doi: 10.1016/j.tim.2004.08.008. Trends Microbiol. 2004. PMID: 15381196 Free PMC article. Review.
Cited by
-
COVID-19 and Carcinogenesis: Exploring the Hidden Links.Cureus. 2024 Aug 31;16(8):e68303. doi: 10.7759/cureus.68303. eCollection 2024 Aug. Cureus. 2024. PMID: 39350850 Free PMC article. Review.
-
Covid-19-induced pulmonary hypertension in children, and the use of phosphodiesterase-5 inhibitors.F1000Res. 2022 Sep 12;10:792. doi: 10.12688/f1000research.53966.2. eCollection 2021. F1000Res. 2022. PMID: 39228925 Free PMC article. Review.
-
Design and Application of Biosafe Coronavirus Engineering Systems without Virulence.Viruses. 2024 Apr 24;16(5):659. doi: 10.3390/v16050659. Viruses. 2024. PMID: 38793541 Free PMC article. Review.
-
Effect of polyphenols against complications of COVID-19: current evidence and potential efficacy.Pharmacol Rep. 2024 Apr;76(2):307-327. doi: 10.1007/s43440-024-00585-6. Epub 2024 Mar 18. Pharmacol Rep. 2024. PMID: 38498260 Review.
-
A Suitable Membrane Distance Regulated by the RBD_ACE2 Interaction is Critical for SARS-CoV-2 Spike-Mediated Viral Invasion.Adv Sci (Weinh). 2023 Oct;10(28):e2301478. doi: 10.1002/advs.202301478. Epub 2023 Aug 17. Adv Sci (Weinh). 2023. PMID: 37590389 Free PMC article.
References
-
- Lee, N., Hui, D., Wu, A., Chan, P., Cameron, P., Joynt, G. M., Ahuja, A., Yung, M. Y., Leung, C. B., To, K. F., et al. (2003) N. Engl. J. Med. 348, 1986-1994. - PubMed
-
- Poutanen, S. M., Low, D. E., Henry, B., Finkelstein, S., Rose, D., Green, K., Tellier, R., Draker, R., Adachi, D., Ayers, M., et al. (2003) N. Engl. J. Med. 348, 1995-2005. - PubMed
-
- Ksiazek, T. G., Erdman, D., Goldsmith, C. S., Zaki, S. R., Peret, T., Emery, S., Tong, S., Urbani, C., Comer, J. A., Lim, W., et al. (2003) N. Engl. J. Med. 348, 1953-1966. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

