The cohesion protein ORD is required for homologue bias during meiotic recombination
- PMID: 15007062
- PMCID: PMC2172286
- DOI: 10.1083/jcb.200310077
The cohesion protein ORD is required for homologue bias during meiotic recombination
Abstract
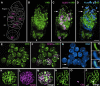
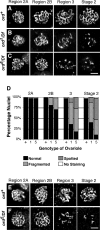
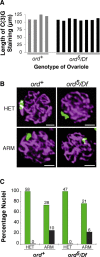
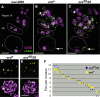
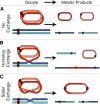
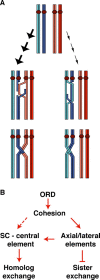
During meiosis, sister chromatid cohesion is required for normal levels of homologous recombination, although how cohesion regulates exchange is not understood. Null mutations in orientation disruptor (ord) ablate arm and centromeric cohesion during Drosophila meiosis and severely reduce homologous crossovers in mutant oocytes. We show that ORD protein localizes along oocyte chromosomes during the stages in which recombination occurs. Although synaptonemal complex (SC) components initially associate with synapsed homologues in ord mutants, their localization is severely disrupted during pachytene progression, and normal tripartite SC is not visible by electron microscopy. In ord germaria, meiotic double strand breaks appear and disappear with frequency and timing indistinguishable from wild type. However, Ring chromosome recovery is dramatically reduced in ord oocytes compared with wild type, which is consistent with the model that defects in meiotic cohesion remove the constraints that normally limit recombination between sisters. We conclude that ORD activity suppresses sister chromatid exchange and stimulates inter-homologue crossovers, thereby promoting homologue bias during meiotic recombination in Drosophila.
Figures

Similar articles
-
Regulation of meiotic cohesion and chromosome core morphogenesis during pachytene in Drosophila oocytes.J Cell Sci. 2007 Sep 1;120(Pt 17):3123-37. doi: 10.1242/jcs.009977. Epub 2007 Aug 14. J Cell Sci. 2007. PMID: 17698920
-
Meiotic cohesion requires accumulation of ORD on chromosomes before condensation.Mol Biol Cell. 2002 Nov;13(11):3890-900. doi: 10.1091/mbc.e02-06-0332. Mol Biol Cell. 2002. PMID: 12429833 Free PMC article.
-
The cohesion protein SOLO associates with SMC1 and is required for synapsis, recombination, homolog bias and cohesion and pairing of centromeres in Drosophila Meiosis.PLoS Genet. 2013;9(7):e1003637. doi: 10.1371/journal.pgen.1003637. Epub 2013 Jul 18. PLoS Genet. 2013. PMID: 23874232 Free PMC article.
-
The molecular control of meiotic chromosomal behavior: events in early meiotic prophase in Drosophila oocytes.Annu Rev Physiol. 2012;74:425-51. doi: 10.1146/annurev-physiol-020911-153342. Annu Rev Physiol. 2012. PMID: 22335798 Review.
-
Prophase I: Preparing Chromosomes for Segregation in the Developing Oocyte.Results Probl Cell Differ. 2017;59:125-173. doi: 10.1007/978-3-319-44820-6_5. Results Probl Cell Differ. 2017. PMID: 28247048 Review.
Cited by
-
Meiosis in mice without a synaptonemal complex.PLoS One. 2011;6(12):e28255. doi: 10.1371/journal.pone.0028255. Epub 2011 Dec 2. PLoS One. 2011. PMID: 22164254 Free PMC article.
-
Pch2 modulates chromatid partner choice during meiotic double-strand break repair in Saccharomyces cerevisiae.Genetics. 2011 Jul;188(3):511-21. doi: 10.1534/genetics.111.129031. Epub 2011 Apr 21. Genetics. 2011. PMID: 21515575 Free PMC article.
-
Dynamic and Stable Cohesins Regulate Synaptonemal Complex Assembly and Chromosome Segregation.Curr Biol. 2016 Jul 11;26(13):1688-1698. doi: 10.1016/j.cub.2016.05.006. Epub 2016 Jun 9. Curr Biol. 2016. PMID: 27291057 Free PMC article.
-
Structural maintenance of chromosomes (SMC) proteins promote homolog-independent recombination repair in meiosis crucial for germ cell genomic stability.PLoS Genet. 2010 Jul 22;6(7):e1001028. doi: 10.1371/journal.pgen.1001028. PLoS Genet. 2010. PMID: 20661436 Free PMC article.
-
All paired up with no place to go: pairing, synapsis, and DSB formation in a balancer heterozygote.PLoS Genet. 2005 Nov;1(5):e67. doi: 10.1371/journal.pgen.0010067. Epub 2005 Nov 18. PLoS Genet. 2005. PMID: 16299588 Free PMC article.
References
-
- Bickel, S.E., T. Orr-Weaver, and E.M. Balicky. 2002. The sister-chromatid cohesion protein ORD is required for chiasma maintenance in Drosophila oocytes. Curr. Biol. 12:925–929. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

