Frequent simian foamy virus infection in persons occupationally exposed to nonhuman primates
- PMID: 14990698
- PMCID: PMC353775
- DOI: 10.1128/jvi.78.6.2780-2789.2004
Frequent simian foamy virus infection in persons occupationally exposed to nonhuman primates
Abstract
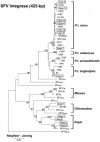
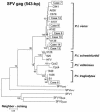
The recognition that AIDS originated as a zoonosis heightens public health concerns associated with human infection by simian retroviruses endemic in nonhuman primates (NHPs). These retroviruses include simian immunodeficiency virus (SIV), simian T-cell lymphotropic virus (STLV), simian type D retrovirus (SRV), and simian foamy virus (SFV). Although occasional infection with SIV, SRV, or SFV in persons occupationally exposed to NHPs has been reported, the characteristics and significance of these zoonotic infections are not fully defined. Surveillance for simian retroviruses at three research centers and two zoos identified no SIV, SRV, or STLV infection in 187 participants. However, 10 of 187 persons (5.3%) tested positive for SFV antibodies by Western blot (WB) analysis. Eight of the 10 were males, and 3 of the 10 worked at zoos. SFV integrase gene (int) and gag sequences were PCR amplified from the peripheral blood lymphocytes available from 9 of the 10 persons. Phylogenetic analysis showed SFV infection originating from chimpanzees (n = 8) and baboons (n = 1). SFV seropositivity for periods of 8 to 26 years (median, 22 years) was documented for six workers for whom archived serum samples were available, demonstrating long-standing SFV infection. All 10 persons reported general good health, and secondary transmission of SFV was not observed in three wives available for WB and PCR testing. Additional phylogenetic analysis of int and gag sequences provided the first direct evidence identifying the source chimpanzees of the SFV infection in two workers. This study documents more frequent infection with SFV than with other simian retroviruses in persons working with NHPs and provides important information on the natural history and species origin of these infections. Our data highlight the importance of studies to better define the public health implications of zoonotic SFV infections.
Figures

Similar articles
-
Detection and molecular characterization of foamy viruses in Central African chimpanzees of the Pan troglodytes troglodytes and Pan troglodytes vellerosus subspecies.J Med Primatol. 2006 Apr;35(2):59-66. doi: 10.1111/j.1600-0684.2006.00149.x. J Med Primatol. 2006. PMID: 16556292
-
Screening for simian foamy virus infection by using a combined antigen Western blot assay: evidence for a wide distribution among Old World primates and identification of four new divergent viruses.Virology. 2003 May 10;309(2):248-57. doi: 10.1016/s0042-6822(03)00070-9. Virology. 2003. PMID: 12758172
-
An Immunodominant and Conserved B-Cell Epitope in the Envelope of Simian Foamy Virus Recognized by Humans Infected with Zoonotic Strains from Apes.J Virol. 2019 May 15;93(11):e00068-19. doi: 10.1128/JVI.00068-19. Print 2019 Jun 1. J Virol. 2019. PMID: 30894477 Free PMC article.
-
Human infection with foamy viruses.Curr Top Microbiol Immunol. 2003;277:181-96. doi: 10.1007/978-3-642-55701-9_8. Curr Top Microbiol Immunol. 2003. PMID: 12908773 Review.
-
Simian retroviruses in African apes.Clin Microbiol Infect. 2012 Jun;18(6):514-20. doi: 10.1111/j.1469-0691.2012.03843.x. Epub 2012 Apr 20. Clin Microbiol Infect. 2012. PMID: 22515409 Review.
Cited by
-
The human-primate interface in the New Normal: Challenges and opportunities for primatologists in the COVID-19 era and beyond.Am J Primatol. 2020 Aug;82(8):e23176. doi: 10.1002/ajp.23176. Epub 2020 Jul 20. Am J Primatol. 2020. PMID: 32686188 Free PMC article.
-
Lentivirus restriction by diverse primate APOBEC3A proteins.Virology. 2013 Jul 20;442(1):82-96. doi: 10.1016/j.virol.2013.04.002. Epub 2013 May 4. Virology. 2013. PMID: 23648232 Free PMC article.
-
Genetic characterization of simian foamy viruses infecting humans.J Virol. 2012 Dec;86(24):13350-9. doi: 10.1128/JVI.01715-12. Epub 2012 Sep 26. J Virol. 2012. PMID: 23015714 Free PMC article.
-
Simian Foamy Viruses in Central and South America: A New World of Discovery.Viruses. 2019 Oct 20;11(10):967. doi: 10.3390/v11100967. Viruses. 2019. PMID: 31635161 Free PMC article. Review.
-
Frequent and recent human acquisition of simian foamy viruses through apes' bites in central Africa.PLoS Pathog. 2011 Oct;7(10):e1002306. doi: 10.1371/journal.ppat.1002306. Epub 2011 Oct 27. PLoS Pathog. 2011. PMID: 22046126 Free PMC article.
References
-
- Achong, B. G., P. W. A. Mansell, M. A. Epstein, and P. J. Clifford. 1971. An unusual virus in cultures from a human nasopharyngeal carcinoma. J. Natl. Cancer Inst. 46:299-307. - PubMed
-
- Ali, M., G. P. Taylor, R. J. Pitman, D. Parker, A. Rethwilm, R. Cheingsong-Popov, J. N. Weber, P. D. Bieniasz, J. Bradley, and M. O. McClure. 1996. No evidence of antibody to human foamy virus in widespread human populations. AIDS Res. Hum. Retrovir. 12:1473-1483. - PubMed
-
- Boneva, R. S., A. Grindon, S. Orton, W. M. Switzer, V. Shanmugam, A. Hussain, V. Bhullar, W. Heneine, M. Chamberland, T. M. Folks, and L. Chapman. 2002. Simian foamy virus infection in a blood donor. Transfusion 42:886-891. - PubMed
-
- Brooks, J. I., E. W. Rud, R. G. Pilon, J. M. Smith, W. M. Switzer, and P. A. Sandstrom. 2002. Cross-species retroviral transmission from macaques to human beings. Lancet 360:387-388. - PubMed
-
- Broussard, S. R., A. G. Comuzzie, K. L. Leighton, M. M. Leland, E. M. Whitehead, and J. S. Allan. 1997. Characterization of new simian foamy viruses from African nonhuman primates. Virology 237:349-359. - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

