Whole-genome annotation by using evidence integration in functional-linkage networks
- PMID: 14981259
- PMCID: PMC365715
- DOI: 10.1073/pnas.0307326101
Whole-genome annotation by using evidence integration in functional-linkage networks
Abstract
The advent of high-throughput biology has catalyzed a remarkable improvement in our ability to identify new genes. A large fraction of newly discovered genes have an unknown functional role, particularly when they are specific to a particular lineage or organism. These genes, currently labeled "hypothetical," might support important biological cell functions and could potentially serve as targets for medical, diagnostic, or pharmacogenomic studies. An important challenge to the scientific community is to associate these newly predicted genes with a biological function that can be validated by experimental screens. In the absence of sequence or structural homology to known genes, we must rely on advanced biotechnological methods, such as DNA chips and protein-protein interaction screens as well as computational techniques to assign putative functions to these genes. In this article, we propose an effective methodology for combining biological evidence obtained in several high-throughput experimental screens and integrating this evidence in a way that provides consistent functional assignments to hypothetical genes. We use the visualization method of propagation diagrams to illustrate the flow of functional evidence that supports the functional assignments produced by the algorithm. Our results contain a number of predictions and furnish strong evidence that integration of functional information is indeed a promising direction for improving the accuracy and robustness of functional genomics.
Figures
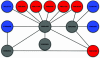
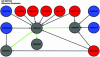
Similar articles
-
AVID: an integrative framework for discovering functional relationships among proteins.BMC Bioinformatics. 2005 Jun 1;6:136. doi: 10.1186/1471-2105-6-136. BMC Bioinformatics. 2005. PMID: 15929793 Free PMC article.
-
Array-based synthetic genetic screens to map bacterial pathways and functional networks in Escherichia coli.Methods Mol Biol. 2011;765:125-53. doi: 10.1007/978-1-61779-197-0_9. Methods Mol Biol. 2011. PMID: 21815091
-
MILANO--custom annotation of microarray results using automatic literature searches.BMC Bioinformatics. 2005 Jan 20;6:12. doi: 10.1186/1471-2105-6-12. BMC Bioinformatics. 2005. PMID: 15661078 Free PMC article.
-
[Transcriptomes for serial analysis of gene expression].J Soc Biol. 2002;196(4):303-7. J Soc Biol. 2002. PMID: 12645300 Review. French.
-
Where are we in genomics?J Physiol Pharmacol. 2005 Jun;56 Suppl 3:37-70. J Physiol Pharmacol. 2005. PMID: 16077195 Review.
Cited by
-
PFP-GO: Integrating protein sequence, domain and protein-protein interaction information for protein function prediction using ranked GO terms.Front Genet. 2022 Sep 29;13:969915. doi: 10.3389/fgene.2022.969915. eCollection 2022. Front Genet. 2022. PMID: 36246645 Free PMC article.
-
Reverse enGENEering of Regulatory Networks from Big Data: A Roadmap for Biologists.Bioinform Biol Insights. 2015 Apr 29;9:61-74. doi: 10.4137/BBI.S12467. eCollection 2015. Bioinform Biol Insights. 2015. PMID: 25983554 Free PMC article. Review.
-
On the detection of functionally coherent groups of protein domains with an extension to protein annotation.BMC Bioinformatics. 2007 Oct 16;8:390. doi: 10.1186/1471-2105-8-390. BMC Bioinformatics. 2007. PMID: 17937820 Free PMC article.
-
VIRGO: computational prediction of gene functions.Nucleic Acids Res. 2006 Jul 1;34(Web Server issue):W340-4. doi: 10.1093/nar/gkl225. Nucleic Acids Res. 2006. PMID: 16845022 Free PMC article.
-
Validation and functional annotation of expression-based clusters based on gene ontology.BMC Bioinformatics. 2006 Aug 15;7:380. doi: 10.1186/1471-2105-7-380. BMC Bioinformatics. 2006. PMID: 16911788 Free PMC article.
References
-
- Gardner, M. J., Shallom, S. J., Carlton, J. M., Salzberg, S. L., Nene, V., Shoaibi, A., Ciecko, A., Lynn, J., Rizzo, M., Weaver, B., et al. (2002) Nature 419, 531-534. - PubMed
-
- Yanai, I., Mellor, J. C. & DeLisi, C. (2002) Trends Genet. 18, 176-179. - PubMed
-
- Marcotte, E. M., Pellegrini, M., Thompson, M. J., Yeates, T. O. & Eisenberg, D. (1999) Nature 402, 83-86. - PubMed
-
- Fields, S. & Song, O. (1989) Nature 340, 245-246. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

