Ordered conformational changes in damaged DNA induced by nucleotide excision repair factors
- PMID: 14981083
- PMCID: PMC4494833
- DOI: 10.1074/jbc.M312611200
Ordered conformational changes in damaged DNA induced by nucleotide excision repair factors
Abstract
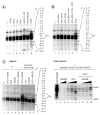
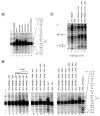
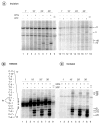
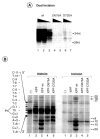
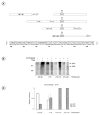
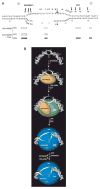
In response to genotoxic attacks, cells activate sophisticated DNA repair pathways such as nucleotide excision repair (NER), which consists of damage removal via dual incision and DNA resynthesis. Using permanganate footprinting as well as highly purified factors, we show that NER is a dynamic process that takes place in a number of successive steps during which the DNA is remodeled around the lesion in response to the various NER factors. XPC/HR23B first recognizes the damaged structure and initiates the opening of the helix from position -3 to +6. TFIIH is then recruited and, in the presence of ATP, extends the opening from position -6 to +6; it also displaces XPC downstream from the lesion, thereby providing the topological structure for recruiting XPA and RPA, which will enlarge the opening. Once targeted by XPG, the damaged DNA is further melted from position -19 to +8. XPG and XPF/ERCC1 endonucleases then cut the damaged DNA at the limit of the opened structure that was previously "labeled" by the positioning of XPC/HR23B and TFIIH.
Figures

Similar articles
-
Mechanism of open complex and dual incision formation by human nucleotide excision repair factors.EMBO J. 1997 Nov 3;16(21):6559-73. doi: 10.1093/emboj/16.21.6559. EMBO J. 1997. PMID: 9351836 Free PMC article.
-
Strong functional interactions of TFIIH with XPC and XPG in human DNA nucleotide excision repair, without a preassembled repairosome.Mol Cell Biol. 2001 Apr;21(7):2281-91. doi: 10.1128/MCB.21.7.2281-2291.2001. Mol Cell Biol. 2001. PMID: 11259578 Free PMC article.
-
The comings and goings of nucleotide excision repair factors on damaged DNA.EMBO J. 2003 Oct 1;22(19):5293-303. doi: 10.1093/emboj/cdg489. EMBO J. 2003. PMID: 14517266 Free PMC article.
-
DNA damage recognition during nucleotide excision repair in mammalian cells.Biochimie. 1999 Jan-Feb;81(1-2):39-44. doi: 10.1016/s0300-9084(99)80036-4. Biochimie. 1999. PMID: 10214908 Review.
-
Xeroderma pigmentosum and molecular cloning of DNA repair genes.Anticancer Res. 1996 Mar-Apr;16(2):693-708. Anticancer Res. 1996. PMID: 8687116 Review.
Cited by
-
New synthetic substrates of mammalian nucleotide excision repair system.Nucleic Acids Res. 2013 Jul;41(12):e123. doi: 10.1093/nar/gkt301. Epub 2013 Apr 22. Nucleic Acids Res. 2013. PMID: 23609543 Free PMC article.
-
Hormonal Regulation of the Repair of UV Photoproducts in Melanocytes by the Melanocortin Signaling Axis.Photochem Photobiol. 2017 Jan;93(1):245-258. doi: 10.1111/php.12640. Epub 2016 Nov 17. Photochem Photobiol. 2017. PMID: 27645605 Free PMC article. Review.
-
The XBP-Bax1 helicase-nuclease complex unwinds and cleaves DNA: implications for eukaryal and archaeal nucleotide excision repair.J Biol Chem. 2010 Apr 2;285(14):11013-22. doi: 10.1074/jbc.M109.094763. Epub 2010 Feb 6. J Biol Chem. 2010. PMID: 20139443 Free PMC article.
-
Probing for DNA damage with β-hairpins: similarities in incision efficiencies of bulky DNA adducts by prokaryotic and human nucleotide excision repair systems in vitro.DNA Repair (Amst). 2011 Jul 15;10(7):684-96. doi: 10.1016/j.dnarep.2011.04.020. Epub 2011 Jul 8. DNA Repair (Amst). 2011. PMID: 21741328 Free PMC article.
-
Physiological consequences of defects in ERCC1-XPF DNA repair endonuclease.DNA Repair (Amst). 2011 Jul 15;10(7):781-91. doi: 10.1016/j.dnarep.2011.04.026. Epub 2011 May 25. DNA Repair (Amst). 2011. PMID: 21612988 Free PMC article. Review.
References
-
- Bohr VA, Smith CA, Okumoto DS, Hanawalt PC. Cell. 1985;40:359–369. - PubMed
-
- Hoeijmakers JH. Nature. 2001;411:366–374. - PubMed
-
- Mellon I, Spivak G, Hanawalt PC. Cell. 1987;51:241–249. - PubMed
-
- De Laat WL, Jaspers NG, Hoeijmakers JH. Genes Dev. 1999;13:768–785. - PubMed
-
- Bootsma D, Kraemer KH, Cleaver JE, Hoeijmakers JHJ. In: The Genetic Basis of Human Cancer. Vogelstein B, Kinzler KW, editors. McGraw-Hill Inc; New York: 1998. pp. 245–274.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

