Pre-steady-state kinetics shows differences in processing of various DNA lesions by Escherichia coli formamidopyrimidine-DNA glycosylase
- PMID: 14769949
- PMCID: PMC373384
- DOI: 10.1093/nar/gkh237
Pre-steady-state kinetics shows differences in processing of various DNA lesions by Escherichia coli formamidopyrimidine-DNA glycosylase
Abstract
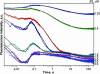
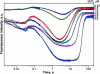
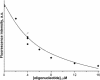
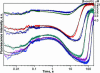
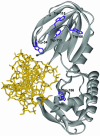
Formamidopyrimidine-DNA-glycosylase (Fpg protein, MutM) catalyses excision of 8-oxoguanine (8-oxoG) and other oxidatively damaged purines from DNA in a glycosylase/apurinic/apyrimidinic-lyase reaction. We report pre-steady-state kinetic analysis of Fpg action on oligonucleotide duplexes containing 8-oxo-2'-deoxyguanosine, natural abasic site or tetrahydrofuran (an uncleavable abasic site analogue). Monitoring Fpg intrinsic tryptophan fluorescence in stopped-flow experiments reveals multiple conformational transitions in the protein molecule during the catalytic cycle. At least four and five conformational transitions occur in Fpg during the interaction with abasic and 8-oxoG-containing substrates, respectively, within 2 ms to 10 s time range. These transitions reflect the stages of enzyme binding to DNA and lesion recognition with the mutual adjustment of DNA and enzyme structures to achieve catalytically competent conformation. Unlike these well-defined binding steps, catalytic stages are not associated with discernible fluorescence events. Only a single conformational change is detected for the cleavable substrates at times exceeding 10 s. The data obtained provide evidence that several fast sequential conformational changes occur in Fpg after binding to its substrate, converting the protein into a catalytically active conformation.
Figures


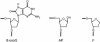
Similar articles
-
Pre-steady-state kinetic study of substrate specificity of Escherichia coli formamidopyrimidine--DNA glycosylase.Biochemistry. 2007 Jan 16;46(2):424-35. doi: 10.1021/bi060787r. Biochemistry. 2007. PMID: 17209553
-
Solution-state NMR investigation of DNA binding interactions in Escherichia coli formamidopyrimidine-DNA glycosylase (Fpg): a dynamic description of the DNA/protein interface.DNA Repair (Amst). 2005 Mar 2;4(3):327-39. doi: 10.1016/j.dnarep.2004.09.012. DNA Repair (Amst). 2005. PMID: 15661656
-
Reversible chemical step and rate-limiting enzyme regeneration in the reaction catalyzed by formamidopyrimidine-DNA glycosylase.Biochemistry. 2009 Dec 8;48(48):11335-43. doi: 10.1021/bi901100b. Biochemistry. 2009. PMID: 19835417
-
Real-time studies of conformational dynamics of the repair enzyme E. coli formamidopyrimidine-DNA glycosylase and its DNA complexes during catalytic cycle.Mutat Res. 2010 Mar 1;685(1-2):3-10. doi: 10.1016/j.mrfmmm.2009.08.018. Epub 2009 Sep 12. Mutat Res. 2010. PMID: 19751748 Review.
-
Recognition of damaged DNA by Escherichia coli Fpg protein: insights from structural and kinetic data.Mutat Res. 2003 Oct 29;531(1-2):141-56. doi: 10.1016/j.mrfmmm.2003.09.002. Mutat Res. 2003. PMID: 14637251 Review.
Cited by
-
Kinetics of substrate recognition and cleavage by human 8-oxoguanine-DNA glycosylase.Nucleic Acids Res. 2005 Jul 15;33(12):3919-31. doi: 10.1093/nar/gki694. Print 2005. Nucleic Acids Res. 2005. PMID: 16024742 Free PMC article.
-
Thermodynamics of the multi-stage DNA lesion recognition and repair by formamidopyrimidine-DNA glycosylase using pyrrolocytosine fluorescence--stopped-flow pre-steady-state kinetics.Nucleic Acids Res. 2012 Aug;40(15):7384-92. doi: 10.1093/nar/gks423. Epub 2012 May 14. Nucleic Acids Res. 2012. PMID: 22584623 Free PMC article.
-
Unusual structural features of hydantoin lesions translate into efficient recognition by Escherichia coli Fpg.Biochemistry. 2007 Aug 21;46(33):9355-65. doi: 10.1021/bi602459v. Epub 2007 Jul 27. Biochemistry. 2007. PMID: 17655276 Free PMC article.
-
Kinetic study of DNA modification by phthalocyanine derivative of the oligonucleotide.Bioinorg Chem Appl. 2006;2006:23560. doi: 10.1155/BCA/2006/23560. Epub 2006 Dec 18. Bioinorg Chem Appl. 2006. PMID: 17497004 Free PMC article.
-
Two glycosylase families diffusively scan DNA using a wedge residue to probe for and identify oxidatively damaged bases.Proc Natl Acad Sci U S A. 2014 May 20;111(20):E2091-9. doi: 10.1073/pnas.1400386111. Epub 2014 May 5. Proc Natl Acad Sci U S A. 2014. PMID: 24799677 Free PMC article.
References
-
- von Sonntag C. (1987) The Chemical Basis of Radiation Biology. Taylor & Francis, London.
-
- Halliwell B. and Gutteridge,J.M.C. (1999) Free Radicals in Biology and Medicine, 3rd Edn. Oxford University Press, Oxford, UK.
-
- Shibutani S., Takeshita,M. and Grollman,A.P. (1991) Insertion of specific bases during DNA synthesis past the oxidation-damaged base 8-oxodG. Nature, 349, 431–434. - PubMed
-
- Moriya M., Ou,C., Bodepudi,V., Johnson,F., Takeshita,M. and Grollman,A.P. (1991) Site-specific mutagenesis using a gapped duplex vector: a study of translesion synthesis past 8-oxodeoxyguanosine in E. coli. Mutat. Res., 254, 281–288. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

