Predicting specificity in bZIP coiled-coil protein interactions
- PMID: 14759261
- PMCID: PMC395749
- DOI: 10.1186/gb-2004-5-2-r11
Predicting specificity in bZIP coiled-coil protein interactions
Abstract
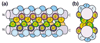
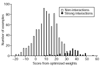
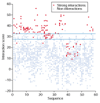
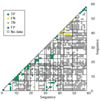
We present a method for predicting protein-protein interactions mediated by the coiled-coil motif. When tested on interactions between nearly all human and yeast bZIP proteins, our method identifies 70% of strong interactions while maintaining that 92% of predictions are correct. Furthermore, cross-validation testing shows that including the bZIP experimental data significantly improves performance. Our method can be used to predict bZIP interactions in other genomes and is a promising approach for predicting coiled-coil interactions more generally.
Figures


Similar articles
-
Structural basis for the diversity of DNA recognition by bZIP transcription factors.Nat Struct Biol. 2000 Oct;7(10):889-93. doi: 10.1038/82822. Nat Struct Biol. 2000. PMID: 11017199
-
Protein-directed DNA structure II. Raman spectroscopy of a leucine zipper bZIP complex.Biochemistry. 2000 Jan 25;39(3):548-56. doi: 10.1021/bi990053x. Biochemistry. 2000. PMID: 10642179
-
Comprehensive identification of human bZIP interactions with coiled-coil arrays.Science. 2003 Jun 27;300(5628):2097-101. doi: 10.1126/science.1084648. Epub 2003 Jun 12. Science. 2003. PMID: 12805554
-
Pharmacological interference with protein-protein interactions mediated by coiled-coil motifs.Handb Exp Pharmacol. 2008;(186):461-82. doi: 10.1007/978-3-540-72843-6_19. Handb Exp Pharmacol. 2008. PMID: 18491064 Review.
-
Interactions of coiled coils in transcription factors: where is the specificity?Curr Opin Genet Dev. 1993 Apr;3(2):278-85. doi: 10.1016/0959-437x(93)90035-n. Curr Opin Genet Dev. 1993. PMID: 8504253 Review.
Cited by
-
Combinatorial control of Arabidopsis proline dehydrogenase transcription by specific heterodimerisation of bZIP transcription factors.EMBO J. 2006 Jul 12;25(13):3133-43. doi: 10.1038/sj.emboj.7601206. Epub 2006 Jun 29. EMBO J. 2006. PMID: 16810321 Free PMC article.
-
Alpha-helical peptide assemblies giving new function to designed structures.Prog Mol Biol Transl Sci. 2011;103:231-75. doi: 10.1016/B978-0-12-415906-8.00001-7. Prog Mol Biol Transl Sci. 2011. PMID: 21999998 Free PMC article. Review.
-
Designing specific protein-protein interactions using computation, experimental library screening, or integrated methods.Protein Sci. 2012 Jul;21(7):949-63. doi: 10.1002/pro.2096. Epub 2012 Jun 8. Protein Sci. 2012. PMID: 22593041 Free PMC article. Review.
-
The genetic architecture of protein interaction affinity and specificity.Nat Commun. 2024 Oct 14;15(1):8868. doi: 10.1038/s41467-024-53195-4. Nat Commun. 2024. PMID: 39402041 Free PMC article.
-
A conserved leucine zipper-like motif accounts for strong tetramerization capabilities of SEPALLATA-like MADS-domain transcription factors.J Exp Bot. 2018 Apr 9;69(8):1943-1954. doi: 10.1093/jxb/ery063. J Exp Bot. 2018. PMID: 29474620 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

