Histone H3 lysine 9 methylation is required for DNA elimination in developing macronuclei in Tetrahymena
- PMID: 14755052
- PMCID: PMC341817
- DOI: 10.1073/pnas.0305421101
Histone H3 lysine 9 methylation is required for DNA elimination in developing macronuclei in Tetrahymena
Abstract
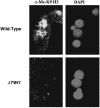
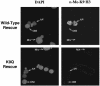
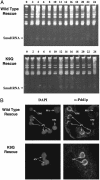
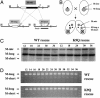
Genome-wide DNA elimination accompanies development of the somatic macronucleus from the germ-line micronucleus during the sexual process of conjugation in the ciliated protozoan Tetrahymena thermophila. Small RNAs, referred to as "scan RNAs" (scnRNAs), that accumulate only during conjugation are highly enriched in the eliminated sequences, and mutations that prevent DNA elimination also affect the accumulation of scnRNAs, suggesting that an RNA interference (RNAi)-like mechanism is involved in DNA elimination. Histone H3 that is methylated at lysine 9 (K9) is a hallmark of heterochromatin and, in Tetrahymena, is found only in developing macronuclei (anlagen) in association with chromatin containing the sequences undergoing elimination. In this article, we demonstrate that a mutation in the TWI1 gene that eliminates the accumulation of scnRNAs also abolishes H3 methylation at K9. We created mutant strains of Tetrahymena in which the only major H3 contained a K9Q mutation. These mutants accumulated scnRNAs normally during conjugation but showed dramatically reduced efficiency of DNA elimination. These results provide strong genetic evidence linking an RNAi-like pathway, H3 K9 methylation, and DNA elimination in Tetrahymena.
Figures

Similar articles
-
Germ line transcripts are processed by a Dicer-like protein that is essential for developmentally programmed genome rearrangements of Tetrahymena thermophila.Mol Cell Biol. 2005 Oct;25(20):9151-64. doi: 10.1128/MCB.25.20.9151-9164.2005. Mol Cell Biol. 2005. PMID: 16199890 Free PMC article.
-
Small RNAs in genome rearrangement in Tetrahymena.Curr Opin Genet Dev. 2004 Apr;14(2):181-7. doi: 10.1016/j.gde.2004.01.004. Curr Opin Genet Dev. 2004. PMID: 15196465 Review.
-
Methylation of histone h3 at lysine 9 targets programmed DNA elimination in tetrahymena.Cell. 2002 Sep 20;110(6):701-11. doi: 10.1016/s0092-8674(02)00941-8. Cell. 2002. PMID: 12297044
-
RNAi-dependent H3K27 methylation is required for heterochromatin formation and DNA elimination in Tetrahymena.Genes Dev. 2007 Jun 15;21(12):1530-45. doi: 10.1101/gad.1544207. Genes Dev. 2007. PMID: 17575054 Free PMC article.
-
Dynamic nuclear reorganization during genome remodeling of Tetrahymena.Biochim Biophys Acta. 2008 Nov;1783(11):2130-6. doi: 10.1016/j.bbamcr.2008.07.012. Epub 2008 Jul 28. Biochim Biophys Acta. 2008. PMID: 18706458 Free PMC article. Review.
Cited by
-
Developmentally programmed, RNA-directed genome rearrangement in Tetrahymena.Dev Growth Differ. 2012 Jan;54(1):108-19. doi: 10.1111/j.1440-169X.2011.01305.x. Epub 2011 Nov 22. Dev Growth Differ. 2012. PMID: 22103557 Free PMC article. Review.
-
A developmentally regulated gene, ASI2, is required for endocycling in the macronuclear anlagen of Tetrahymena.Eukaryot Cell. 2010 Sep;9(9):1343-53. doi: 10.1128/EC.00089-10. Epub 2010 Jul 23. Eukaryot Cell. 2010. PMID: 20656911 Free PMC article.
-
Elimination of foreign DNA during somatic differentiation in Tetrahymena thermophila shows position effect and is dosage dependent.Eukaryot Cell. 2005 Feb;4(2):421-31. doi: 10.1128/EC.4.2.421-431.2005. Eukaryot Cell. 2005. PMID: 15701804 Free PMC article.
-
Small RNAs prevent transcription-coupled loss of histone H3 lysine 9 methylation in Arabidopsis thaliana.PLoS Genet. 2011 Oct;7(10):e1002350. doi: 10.1371/journal.pgen.1002350. Epub 2011 Oct 27. PLoS Genet. 2011. PMID: 22046144 Free PMC article.
-
Uniting germline and stem cells: the function of Piwi proteins and the piRNA pathway in diverse organisms.Annu Rev Genet. 2011;45:447-69. doi: 10.1146/annurev-genet-110410-132541. Epub 2011 Sep 19. Annu Rev Genet. 2011. PMID: 21942366 Free PMC article. Review.
References
-
- Jenuwein, T. & Allis, C. D. (2001) Science 293, 1074-1080. - PubMed
-
- Grewal, S. I. & Elgin, S. C. (2002) Curr. Opin. Genet. Dev. 12, 178-187. - PubMed
-
- Lachner, M. & Jenuwein, T. (2002) Curr. Opin. Cell Biol. 14, 286-298. - PubMed
-
- Bannister, A. J., Zegerman, P., Partridge, J. F., Miska, E. A., Thomas, J. O., Allshire, R. C. & Kouzarides, T. (2001) Nature 410, 120-124. - PubMed
-
- Lachner, M., O'Carroll, D., Rea, S., Mechtler, K. & Jenuwein, T. (2001) Nature 410, 116-120. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

