Molecular clearance of ataxin-3 is regulated by a mammalian E4
- PMID: 14749733
- PMCID: PMC1271811
- DOI: 10.1038/sj.emboj.7600081
Molecular clearance of ataxin-3 is regulated by a mammalian E4
Abstract
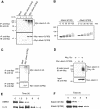
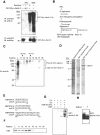
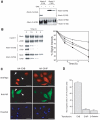
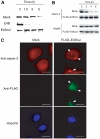
Insoluble aggregates of polyglutamine-containing proteins are usually conjugated with ubiquitin in neurons of individuals with polyglutamine diseases. We now show that ataxin-3, in which the abnormal expansion of a polyglutamine tract is responsible for spinocerebellar ataxia type 3 (SCA3), undergoes ubiquitylation and degradation by the proteasome. Mammalian E4B (UFD2a), a ubiquitin chain assembly factor (E4), copurified with the polyubiquitylation activity for ataxin-3. E4B interacted with, and thereby mediated polyubiquitylation of, ataxin-3. Expression of E4B promoted degradation of a pathological form of ataxin-3. In contrast, a dominant-negative mutant of E4B inhibited degradation of this form of ataxin-3, resulting in the formation of intracellular aggregates. In a Drosophila model of SCA3, expression of E4B suppressed the neurodegeneration induced by an ataxin-3 mutant. These observations suggest that E4 is a rate-limiting factor in the degradation of pathological forms of ataxin-3, and that targeted expression of E4B is a potential gene therapy for SCA3.
Figures


Similar articles
-
[Neurodegenerative diseases regulated by ubiquitin-proteasome system].Rinsho Shinkeigaku. 2006 Nov;46(11):890-2. Rinsho Shinkeigaku. 2006. PMID: 17432211 Japanese.
-
[Mechanisms to control degradation of polyglutamine-containing protein].Rinsho Shinkeigaku. 2003 Nov;43(11):906-8. Rinsho Shinkeigaku. 2003. PMID: 15152500 Review. Japanese.
-
Ataxin-3 suppresses polyglutamine neurodegeneration in Drosophila by a ubiquitin-associated mechanism.Mol Cell. 2005 Apr 1;18(1):37-48. doi: 10.1016/j.molcel.2005.02.030. Mol Cell. 2005. PMID: 15808507
-
Ataxin-3 protein modification as a treatment strategy for spinocerebellar ataxia type 3: removal of the CAG containing exon.Neurobiol Dis. 2013 Oct;58:49-56. doi: 10.1016/j.nbd.2013.04.019. Epub 2013 May 6. Neurobiol Dis. 2013. PMID: 23659897
-
Progress in pathogenesis studies of spinocerebellar ataxia type 1.Philos Trans R Soc Lond B Biol Sci. 1999 Jun 29;354(1386):1079-81. doi: 10.1098/rstb.1999.0462. Philos Trans R Soc Lond B Biol Sci. 1999. PMID: 10434309 Free PMC article. Review.
Cited by
-
Unbiased screen identifies aripiprazole as a modulator of abundance of the polyglutamine disease protein, ataxin-3.Brain. 2016 Nov 1;139(11):2891-2908. doi: 10.1093/brain/aww228. Brain. 2016. PMID: 27645800 Free PMC article.
-
Deubiquitinating function of ataxin-3: insights from the solution structure of the Josephin domain.Proc Natl Acad Sci U S A. 2005 Sep 6;102(36):12700-5. doi: 10.1073/pnas.0506344102. Epub 2005 Aug 23. Proc Natl Acad Sci U S A. 2005. PMID: 16118278 Free PMC article.
-
The ubiquitin-proteasome system.J Biosci. 2006 Mar;31(1):137-55. doi: 10.1007/BF02705243. J Biosci. 2006. PMID: 16595883 Review.
-
In vivo suppression of polyglutamine neurotoxicity by C-terminus of Hsp70-interacting protein (CHIP) supports an aggregation model of pathogenesis.Neurobiol Dis. 2009 Mar;33(3):342-53. doi: 10.1016/j.nbd.2008.10.016. Epub 2008 Nov 8. Neurobiol Dis. 2009. PMID: 19084066 Free PMC article.
-
Identification of an RNA Polymerase III Regulator Linked to Disease-Associated Protein Aggregation.Mol Cell. 2017 Mar 16;65(6):1096-1108.e6. doi: 10.1016/j.molcel.2017.02.022. Mol Cell. 2017. PMID: 28306505 Free PMC article.
References
-
- Alves-Rodrigues A, Gregori L, Figueiredo-Pereira ME (1998) Ubiquitin, cellular inclusions and their role in neurodegeneration. Trends Neurosci 21: 516–520 - PubMed
-
- Aravind L, Koonin EV (2000) The U box is a modified RING finger—a common domain in ubiquitination. Curr Biol 10: R132–R134 - PubMed
-
- Banerjee U, Renfranz PJ, Pollock JA, Benzer S (1987) Molecular characterization and expression of sevenless, a gene involved in neuronal pattern formation in the Drosophila eye. Cell 49: 281–291 - PubMed
-
- Brand AH, Perrimon N (1993) Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118: 401–415 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

