Premature termination codons enhance mRNA decapping in human cells
- PMID: 14742663
- PMCID: PMC373342
- DOI: 10.1093/nar/gkh218
Premature termination codons enhance mRNA decapping in human cells
Abstract
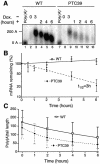
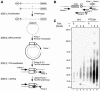
Nonsense-mediated mRNA decay (NMD) is a eukaryotic surveillance process that promotes selective degradation of imperfect messages containing premature translation termination codons (PTCs). In yeast, PTCs trigger both deadenylylation-independent mRNA decapping, thereby allowing their rapid degradation by a 5' to 3' exonuclease, and to a smaller extent accelerated deadenylylation. It is not clear to what extent this decay pathway is conserved in higher eukaryotes. We used a transcriptional pulse strategy relying on a tetracycline-regulated promoter to study the decay of a PTC- containing beta-globin mRNA in human cells. We show that a PTC destabilizes the mRNA and decreases its half-life from >16 h to 3 h. The deadenylylation rate is increased, but not sufficiently to account for the decreased half-life on its own. Using a circularization RT-PCR (cRT-PCR) strategy, we could detect decapped degradation intermediates and measure simultaneously their poly(A) tail length. This allowed us to show that a PTC enhances the rate of mRNA decapping and that decapped products have been deadenylylated to a certain extent. Thus the major feature of the NMD pathway, enhanced decapping, is conserved from yeast to man even though the kinetic details might differ between various mRNAs and/or species.
Figures
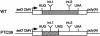
Similar articles
-
[Nonsense-mediated mRNA decay (NMD)--on guard of mRNA quality].Postepy Biochem. 2006;52(4):390-8. Postepy Biochem. 2006. PMID: 17536508 Review. Polish.
-
EJC-independent degradation of nonsense immunoglobulin-mu mRNA depends on 3' UTR length.Nat Struct Mol Biol. 2006 May;13(5):462-4. doi: 10.1038/nsmb1081. Epub 2006 Apr 23. Nat Struct Mol Biol. 2006. PMID: 16622410
-
Premature translational termination triggers mRNA decapping.Nature. 1994 Aug 18;370(6490):578-81. doi: 10.1038/370578a0. Nature. 1994. PMID: 8052314
-
The mammalian nonsense-mediated mRNA decay pathway: to decay or not to decay! Which players make the decision?FEBS Lett. 2009 Feb 4;583(3):499-505. doi: 10.1016/j.febslet.2008.12.058. Epub 2009 Jan 20. FEBS Lett. 2009. PMID: 19162024 Review.
-
Nonsense-mediated mRNA decay: molecular insights and mechanistic variations across species.Curr Opin Cell Biol. 2005 Jun;17(3):316-25. doi: 10.1016/j.ceb.2005.04.005. Curr Opin Cell Biol. 2005. PMID: 15901503 Review.
Cited by
-
A novel frameshift mutation (+G) at codons 15/16 in a beta0 thalassaemia gene results in a significant reduction of beta globin mRNA values.J Clin Pathol. 2005 Sep;58(9):923-6. doi: 10.1136/jcp.2004.025296. J Clin Pathol. 2005. PMID: 16126871 Free PMC article.
-
Cytoplasmic foci are sites of mRNA decay in human cells.J Cell Biol. 2004 Apr;165(1):31-40. doi: 10.1083/jcb.200309008. Epub 2004 Apr 5. J Cell Biol. 2004. PMID: 15067023 Free PMC article.
-
Human nonsense-mediated RNA decay initiates widely by endonucleolysis and targets snoRNA host genes.Genes Dev. 2014 Nov 15;28(22):2498-517. doi: 10.1101/gad.246538.114. Genes Dev. 2014. PMID: 25403180 Free PMC article.
-
A Comparative Overview of the Role of Human Ribonucleases in Nonsense-Mediated mRNA Decay.Genes (Basel). 2024 Oct 10;15(10):1308. doi: 10.3390/genes15101308. Genes (Basel). 2024. PMID: 39457432 Free PMC article.
-
Mechanisms of endonuclease-mediated mRNA decay.Wiley Interdiscip Rev RNA. 2011 Jul-Aug;2(4):582-600. doi: 10.1002/wrna.78. Epub 2011 Feb 10. Wiley Interdiscip Rev RNA. 2011. PMID: 21957046 Free PMC article. Review.
References
-
- Hentze M.W. and Kulozik,A.E. (1999) A perfect message: RNA surveillance and nonsense-mediated decay. Cell, 96, 307–310. - PubMed
-
- Wilusz C.J., Wang,W. and Peltz,S.W. (2001) Curbing the non-sense: the activation and regulation of mRNA surveillance. Genes Dev., 15, 2781–2785. - PubMed
-
- Oliveira C.C. and McCarthy,J.E. (1995) The relationship between eukaryotic translation and mRNA stability. A short upstream open reading frame strongly inhibits translational initiation and greatly accelerates mRNA degradation in the yeast Saccharomyces cerevisiae. J. Biol. Chem., 270, 8936–8943. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

