Role of Saccharomyces single-stranded DNA-binding protein RPA in the strand invasion step of double-strand break repair
- PMID: 14737196
- PMCID: PMC314472
- DOI: 10.1371/journal.pbio.0020021
Role of Saccharomyces single-stranded DNA-binding protein RPA in the strand invasion step of double-strand break repair
Abstract
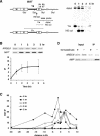
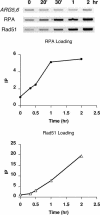
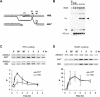
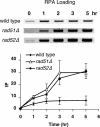
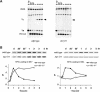
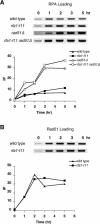
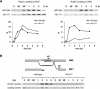
The single-stranded DNA (ssDNA)-binding protein replication protein A (RPA) is essential for both DNA replication and recombination. Chromatin immunoprecipitation techniques were used to visualize the kinetics and extent of RPA binding following induction of a double-strand break (DSB) and during its repair by homologous recombination in yeast. RPA assembles at the HO endonuclease-cut MAT locus simultaneously with the appearance of the DSB, and binding spreads away from the DSB as 5' to 3' exonuclease activity creates more ssDNA. RPA binding precedes binding of the Rad51 recombination protein. The extent of RPA binding is greater when Rad51 is absent, supporting the idea that Rad51 displaces RPA from ssDNA. RPA plays an important role during RAD51-mediated strand invasion of the MAT ssDNA into the donor sequence HML. The replication-proficient but recombination-defective rfa1-t11 (K45E) mutation in the large subunit of RPA is normal in facilitating Rad51 filament formation on ssDNA, but is unable to achieve synapsis between MAT and HML. Thus, RPA appears to play a role in strand invasion as well as in facilitating Rad51 binding to ssDNA, possibly by stabilizing the displaced ssDNA.
Conflict of interest statement
The authors have declared that no conflicts of interest exist.
Figures
Similar articles
-
Dynamic regulatory interactions of rad51, rad52, and replication protein-a in recombination intermediates.J Mol Biol. 2009 Jul 3;390(1):45-55. doi: 10.1016/j.jmb.2009.05.009. Epub 2009 May 13. J Mol Biol. 2009. PMID: 19445949
-
The recombination-deficient mutant RPA (rfa1-t11) is displaced slowly from single-stranded DNA by Rad51 protein.J Biol Chem. 2003 Jun 27;278(26):23410-7. doi: 10.1074/jbc.M302995200. Epub 2003 Apr 14. J Biol Chem. 2003. PMID: 12697761
-
The processing of double-strand breaks and binding of single-strand-binding proteins RPA and Rad51 modulate the formation of ATR-kinase foci in yeast.J Cell Sci. 2007 Dec 1;120(Pt 23):4209-20. doi: 10.1242/jcs.018366. Epub 2007 Nov 14. J Cell Sci. 2007. PMID: 18003698
-
Replication protein A: single-stranded DNA's first responder: dynamic DNA-interactions allow replication protein A to direct single-strand DNA intermediates into different pathways for synthesis or repair.Bioessays. 2014 Dec;36(12):1156-61. doi: 10.1002/bies.201400107. Epub 2014 Aug 29. Bioessays. 2014. PMID: 25171654 Free PMC article. Review.
-
Chaperoning RPA during DNA metabolism.Curr Genet. 2019 Aug;65(4):857-864. doi: 10.1007/s00294-019-00945-3. Epub 2019 Feb 22. Curr Genet. 2019. PMID: 30796471 Review.
Cited by
-
Novel checkpoint response to genotoxic stress mediated by nucleolin-replication protein a complex formation.Mol Cell Biol. 2005 Mar;25(6):2463-74. doi: 10.1128/MCB.25.6.2463-2474.2005. Mol Cell Biol. 2005. PMID: 15743838 Free PMC article.
-
The consequences of asynapsis for mammalian meiosis.Nat Rev Genet. 2009 Mar;10(3):207-16. doi: 10.1038/nrg2505. Nat Rev Genet. 2009. PMID: 19188923 Review.
-
Molecular anatomy of the recombination mediator function of Saccharomyces cerevisiae Rad52.J Biol Chem. 2008 May 2;283(18):12166-74. doi: 10.1074/jbc.M800763200. Epub 2008 Feb 29. J Biol Chem. 2008. PMID: 18310075 Free PMC article.
-
Functional targeting of DNA damage to a nuclear pore-associated SUMO-dependent ubiquitin ligase.Science. 2008 Oct 24;322(5901):597-602. doi: 10.1126/science.1162790. Science. 2008. PMID: 18948542 Free PMC article.
-
Replication protein A prevents promiscuous annealing between short sequence homologies: Implications for genome integrity.Bioessays. 2015 Mar;37(3):305-13. doi: 10.1002/bies.201400161. Epub 2014 Nov 14. Bioessays. 2015. PMID: 25400143 Free PMC article. Review.
References
-
- Alani E, Thresher R, Griffith JD, Kolodner RD. Characterization of DNA-binding and strand-exchange stimulation properties of y-RPA, a yeast single-strand-DNA-binding protein. J Mol Biol. 1992;227:54–71. - PubMed
-
- Aparicio OM, Weinstein DM, Bell SP. Components and dynamics of DNA replication complexes in S. cerevisiae: Redistribution of MCM proteins and Cdc45p during S phase. Cell. 1997;91:59–69. - PubMed
-
- Bianco PR, Tracy RB, Kowalczykowski SC. DNA strand exchange proteins: A biochemical and physical comparison. Front Biosci. 1998;3:D570–D603. - PubMed
-
- Brill SJ, Stillman B. Replication factor-A from Saccharomyces cerevisiae is encoded by three essential genes coordinately expressed at S phase. Genes Dev. 1991;5:1589–1600. - PubMed
-
- Dedon PC, Soults JA, Allis CD, Gorovsky MA. A simplified formaldehyde fixation and immunoprecipitation technique for studying protein–DNA interactions. Anal Biochem. 1991;197:83–90. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

