Characterization of cytosine methylated regions and 5-cytosine DNA methyltransferase (Ehmeth) in the protozoan parasite Entamoeba histolytica
- PMID: 14715927
- PMCID: PMC373271
- DOI: 10.1093/nar/gkh161
Characterization of cytosine methylated regions and 5-cytosine DNA methyltransferase (Ehmeth) in the protozoan parasite Entamoeba histolytica
Abstract
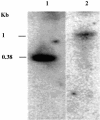
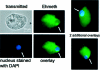
The DNA methylation status of the protozoan parasite Entamoeba histolytica was heretofore unknown. In the present study, we developed a new technique, based on the affinity of methylated DNA to 5-methylcytosine antibodies, to identify methylated DNA in this parasite. Ribosomal DNA and ribosomal DNA circles were isolated by this method and we confirmed the validity of our approach by sodium bisulfite sequencing. We also report the identification and the characterization of a gene, Ehmeth, encoding a DNA methyltransferase strongly homologous to the human DNA methyltransferase 2 (Dnmt2). Immunofluorescence microscopy using an antibody raised against a recombinant Ehmeth showed that Ehmeth is concentrated in the nuclei of trophozoites. The recombinant Ehmeth has a weak but significant methyltransferase activity when E.histolytica genomic DNA is used as substrate. 5-Azacytidine (5-AzaC), an inhibitor of DNA methyltransferase, was used to study in vivo the role of DNA methylation in E.histolytica. Genomic DNA of trophozoites grown with 5-AzaC (23 microM) was undermethylated and the ability of 5-AzaC-treated trophozoites to kill mammalian cells or to cause liver abscess in hamsters was strongly impaired.
Figures






Similar articles
-
Growth of the protozoan parasite Entamoeba histolytica in 5-azacytidine has limited effects on parasite gene expression.BMC Genomics. 2007 Jan 5;8:7. doi: 10.1186/1471-2164-8-7. BMC Genomics. 2007. PMID: 17207281 Free PMC article.
-
Sensing DNA methylation in the protozoan parasite Entamoeba histolytica.Mol Microbiol. 2006 Dec;62(5):1373-86. doi: 10.1111/j.1365-2958.2006.05464.x. Epub 2006 Oct 24. Mol Microbiol. 2006. PMID: 17059565
-
Entamoeba histolytica DNA methyltransferase (Ehmeth) is a nuclear matrix protein that binds EhMRS2, a DNA that includes a scaffold/matrix attachment region (S/MAR).Mol Biochem Parasitol. 2005 Jan;139(1):91-7. doi: 10.1016/j.molbiopara.2004.10.003. Mol Biochem Parasitol. 2005. PMID: 15610823
-
New insights into Entamoeba histolytica pathogenesis.Curr Opin Infect Dis. 2008 Oct;21(5):489-94. doi: 10.1097/QCO.0b013e32830ce75f. Curr Opin Infect Dis. 2008. PMID: 18725798 Free PMC article. Review.
-
Entamoeba histolytica: phylogenetic considerations. A review.Arch Med Res. 1992;23(2):1-5. Arch Med Res. 1992. PMID: 1340267 Review.
Cited by
-
Developmentally regulated DNA methylation in Dictyostelium discoideum.Eukaryot Cell. 2006 Jan;5(1):18-25. doi: 10.1128/EC.5.1.18-25.2006. Eukaryot Cell. 2006. PMID: 16400165 Free PMC article.
-
DNA Methylation of Gene Expression in Acanthamoeba castellanii Encystation.Korean J Parasitol. 2017 Apr;55(2):115-120. doi: 10.3347/kjp.2017.55.2.115. Epub 2017 Apr 30. Korean J Parasitol. 2017. PMID: 28506032 Free PMC article.
-
Transcriptome Analysis of ppdnmt2 and Identification of Superoxide Dismutase as a Novel Interactor of DNMT2 in the Moss Physcomitrella patens.Front Plant Sci. 2020 Aug 5;11:1185. doi: 10.3389/fpls.2020.01185. eCollection 2020. Front Plant Sci. 2020. PMID: 32849734 Free PMC article.
-
A new nuclear function of the Entamoeba histolytica glycolytic enzyme enolase: the metabolic regulation of cytosine-5 methyltransferase 2 (Dnmt2) activity.PLoS Pathog. 2010 Feb 19;6(2):e1000775. doi: 10.1371/journal.ppat.1000775. PLoS Pathog. 2010. PMID: 20174608 Free PMC article.
-
Are Metabolites From the Gut Microbiota Capable of Regulating Epigenetic Mechanisms in the Human Parasite Entamoeba histolytica?Front Cell Dev Biol. 2022 Mar 1;10:841586. doi: 10.3389/fcell.2022.841586. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 35300430 Free PMC article. Review.
References
-
- World Health Organization (1997) Amoebiasis. WHO Weekly Epidemiologic Record, 72, 97–100.
-
- Stanley S.L. (2001) Pathophysiology of amoebiasis. Trends Parasitol., 17, 280–285. - PubMed
-
- Lewin B., Stanley,S.L.,Jr and Reed,S.L. (1998) The mystique of epigenetics. Cell, 93, 301–303. - PubMed
-
- Bestor T.H., Verdine,G.L., Santi,D.V., Norment,A., Garrett,C.E., Gilchrist,C.A., Petri,W.A., Singh,V.K., Moskovitz,J., Wilkinson,B.J. et al. (1994) DNA methyltransferases. Curr. Opin. Cell Biol., 6, 380–389. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

