Stress-induced transcription of satellite III repeats
- PMID: 14699086
- PMCID: PMC2171959
- DOI: 10.1083/jcb.200306104
Stress-induced transcription of satellite III repeats
Abstract
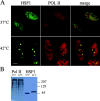
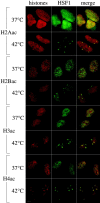
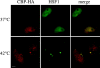
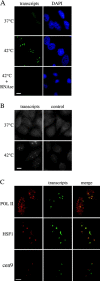
Exposure of mammalian cells to stress induces the activation of heat shock transcription factor 1 (HSF1) and the subsequent transcription of heat shock genes. Activation of the heat shock response also correlates with a rapid relocalization of HSF1 within a few nuclear structures termed nuclear stress granules. These stress-induced structures, which form primarily on the 9q12 region in humans through direct binding of HSF1 to satellite III repeats, do not colocalize with transcription sites of known hsp genes. In this paper, we show that nuclear stress granules correspond to RNA polymerase II transcription factories where satellite III repeats are transcribed into large and stable RNAs that remain associated with the 9q12 region, even throughout mitosis. This work not only reveals the existence of a new major heat-induced transcript in human cells that may play a role in chromatin structure, but also provides evidence for a transcriptional activity within a locus considered so far as heterochromatic and silent.
Figures

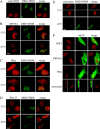


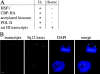
Comment in
-
Nuclear stress granules: the awakening of a sleeping beauty?J Cell Biol. 2004 Jan 5;164(1):15-7. doi: 10.1083/jcb.200311102. J Cell Biol. 2004. PMID: 14709538 Free PMC article. Review.
Similar articles
-
A key role for stress-induced satellite III transcripts in the relocalization of splicing factors into nuclear stress granules.J Cell Sci. 2004 Sep 1;117(Pt 19):4551-8. doi: 10.1242/jcs.01329. J Cell Sci. 2004. PMID: 15331664
-
Nuclear stress granules: the awakening of a sleeping beauty?J Cell Biol. 2004 Jan 5;164(1):15-7. doi: 10.1083/jcb.200311102. J Cell Biol. 2004. PMID: 14709538 Free PMC article. Review.
-
In vivo binding of active heat shock transcription factor 1 to human chromosome 9 heterochromatin during stress.J Cell Biol. 2002 Mar 4;156(5):775-81. doi: 10.1083/jcb.200109018. Epub 2002 Mar 4. J Cell Biol. 2002. PMID: 11877455 Free PMC article.
-
Heat shock factor 1 binds to and transcribes satellite II and III sequences at several pericentromeric regions in heat-shocked cells.Exp Cell Res. 2010 Jul 1;316(11):1845-55. doi: 10.1016/j.yexcr.2010.02.002. Epub 2010 Feb 10. Exp Cell Res. 2010. PMID: 20152833
-
Satellite DNA-mediated effects on genome regulation.Genome Dyn. 2012;7:153-69. doi: 10.1159/000337116. Epub 2012 Jun 25. Genome Dyn. 2012. PMID: 22759818 Review.
Cited by
-
Caenorhabditis elegans HSF-1 is an essential nuclear protein that forms stress granule-like structures following heat shock.Aging Cell. 2013 Feb;12(1):112-20. doi: 10.1111/acel.12024. Epub 2012 Nov 23. Aging Cell. 2013. PMID: 23107491 Free PMC article.
-
Combined Assay of rDNA and SatIII Copy Numbers as an Individual Profile of Stress Resistance, Longevity, Fertility and Disease Predisposition.J Pers Med. 2022 Oct 21;12(10):1752. doi: 10.3390/jpm12101752. J Pers Med. 2022. PMID: 36294891 Free PMC article. Review.
-
Transcribed Tc1-like transposons in salmonid fish.BMC Genomics. 2005 Aug 12;6:107. doi: 10.1186/1471-2164-6-107. BMC Genomics. 2005. PMID: 16095544 Free PMC article.
-
Human sat III and Drosophila hsr omega transcripts: a common paradigm for regulation of nuclear RNA processing in stressed cells.Nucleic Acids Res. 2006;34(19):5508-14. doi: 10.1093/nar/gkl711. Epub 2006 Oct 4. Nucleic Acids Res. 2006. PMID: 17020918 Free PMC article. Review.
-
Mutant nuclear lamin A leads to progressive alterations of epigenetic control in premature aging.Proc Natl Acad Sci U S A. 2006 Jun 6;103(23):8703-8. doi: 10.1073/pnas.0602569103. Epub 2006 May 31. Proc Natl Acad Sci U S A. 2006. PMID: 16738054 Free PMC article.
References
-
- Avner, P., and E. Heard. 2001. X-chromosome inactivation: counting, choice and initiation. Nat. Rev. Genet. 2:59–67. - PubMed
-
- Chiodi, I., M. Biggiogera, M. Denegri, M. Corioni, F. Weighardt, F. Cobianchi, S. Riva, and G. Biamonti. 2000. Structure and dynamics of hnRNP-labelled nuclear bodies induced by stress treatments. J. Cell Sci. 113:4043–4053. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

