5'- and 3'-noncoding regions in flavivirus RNA
- PMID: 14696330
- PMCID: PMC7119107
- DOI: 10.1016/s0065-3527(03)59006-6
5'- and 3'-noncoding regions in flavivirus RNA
Abstract
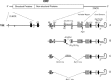
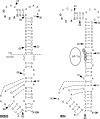
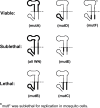
The flavivirus genome is a capped, positive-sense RNA approximately 10.5 kb in length. It contains a single long open reading frame (ORF), flanked by a 5´ noncoding regions (NCR), which is about 100 nucleotides in length, and a 3´ NCR ranging in size from about 400 to 800 nucleotides in length. The conserved structural and nucleotide sequence elements of these NCRs and their function in RNA replication and translation are the subjects of this chapter. The 5´ and 3´ NCRs play a role in the initiation of negative-strand synthesis on virus RNA released from entering virions, switching from negative-strand synthesis to synthesis of progeny plus strand RNA at late times after infection, and possibly in the initiation of translation and in the packaging of virus plus strand RNA into particles. The presence of conserved and nonconserved complementary nucleotide sequences near the 5´ and 3´ termini of flavivirus genomes suggests that ‘‘panhandle’’ or circular RNA structures are formed transiently by hydrogen bonding at some stage during RNA replication.
Figures





Similar articles
-
[Structure and function of HAV genome (RNA)].Nihon Rinsho. 2004 Aug;62 Suppl 8:428-32. Nihon Rinsho. 2004. PMID: 15453360 Review. Japanese. No abstract available.
-
Inhibition of flavivirus infections by antisense oligomers specifically suppressing viral translation and RNA replication.J Virol. 2005 Apr;79(8):4599-609. doi: 10.1128/JVI.79.8.4599-4609.2005. J Virol. 2005. PMID: 15795246 Free PMC article.
-
The 5' and 3' downstream AUG region elements are required for mosquito-borne flavivirus RNA replication.J Virol. 2011 Feb;85(4):1900-5. doi: 10.1128/JVI.02037-10. Epub 2010 Dec 1. J Virol. 2011. PMID: 21123391 Free PMC article.
-
[Structure and function of the non-coding regions of hepatitis C viral RNA].Postepy Biochem. 2006;52(1):62-71. Postepy Biochem. 2006. PMID: 16869303 Review. Polish.
-
In vitro RNA synthesis from exogenous dengue viral RNA templates requires long range interactions between 5'- and 3'-terminal regions that influence RNA structure.J Biol Chem. 2001 May 11;276(19):15581-91. doi: 10.1074/jbc.M010923200. Epub 2001 Feb 5. J Biol Chem. 2001. PMID: 11278787
Cited by
-
Information Encoded by the Flavivirus Genomes beyond the Nucleotide Sequence.Int J Mol Sci. 2021 Apr 3;22(7):3738. doi: 10.3390/ijms22073738. Int J Mol Sci. 2021. PMID: 33916729 Free PMC article. Review.
-
Molecular Insights into the Flavivirus Replication Complex.Viruses. 2021 May 21;13(6):956. doi: 10.3390/v13060956. Viruses. 2021. PMID: 34064113 Free PMC article. Review.
-
Genetic variation in the 3' untranslated region of dengue virus serotype 3 strains isolated from mosquitoes and humans in Brazil.Virol J. 2013 Jan 2;10:3. doi: 10.1186/1743-422X-10-3. Virol J. 2013. PMID: 23282086 Free PMC article.
-
RNA sequences and structures required for the recruitment and activity of the dengue virus polymerase.J Biol Chem. 2011 Mar 4;286(9):6929-39. doi: 10.1074/jbc.M110.162289. Epub 2010 Dec 23. J Biol Chem. 2011. PMID: 21183683 Free PMC article.
-
Molecular Characterization of Dengue Virus Serotype 2 Cosmospolitan Genotype From 2015 Dengue Outbreak in Yunnan, China.Front Cell Infect Microbiol. 2018 Jun 27;8:219. doi: 10.3389/fcimb.2018.00219. eCollection 2018. Front Cell Infect Microbiol. 2018. PMID: 29998087 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
