Sequence motifs involved in the regulation of discontinuous coronavirus subgenomic RNA synthesis
- PMID: 14694129
- PMCID: PMC368802
- DOI: 10.1128/jvi.78.2.980-994.2004
Sequence motifs involved in the regulation of discontinuous coronavirus subgenomic RNA synthesis
Abstract
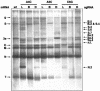
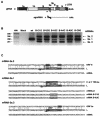
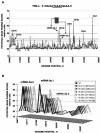
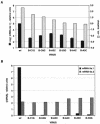
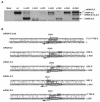
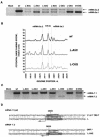
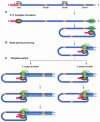
Coronavirus transcription leads to the synthesis of a nested set of mRNAs with a leader sequence derived from the 5' end of the genome. The mRNAs are produced by a discontinuous transcription in which the leader is linked to the mRNA coding sequences. This process is regulated by transcription-regulating sequences (TRSs) preceding each mRNA, including a highly conserved core sequence (CS) with high identity to sequences present in the virus genome and at the 3' end of the leader (TRS-L). The role of TRSs was analyzed by reverse genetics using a full-length infectious coronavirus cDNA and site-directed mutagenesis of the CS. The canonical CS-B was nonessential for the generation of subgenomic mRNAs (sgmRNAs), but its presence led to transcription levels at least 10(3)-fold higher than those in its absence. The data obtained are compatible with a transcription mechanism including three steps: (i) formation of 5'-3' complexes in the genomic RNA, (ii) base-pairing scanning of the nascent negative RNA strand by the TRS-L, and (iii) template switching during synthesis of the negative strand to complete the negative sgRNA. This template switch takes place after copying the CS sequence and was predicted in silico based on high base-pairing score between the nascent negative RNA strand and the TRS-L and minimum DeltaG.
Figures




Similar articles
-
Role of nucleotides immediately flanking the transcription-regulating sequence core in coronavirus subgenomic mRNA synthesis.J Virol. 2005 Feb;79(4):2506-16. doi: 10.1128/JVI.79.4.2506-2516.2005. J Virol. 2005. PMID: 15681451 Free PMC article.
-
Identification of a coronavirus transcription enhancer.J Virol. 2008 Apr;82(8):3882-93. doi: 10.1128/JVI.02622-07. Epub 2008 Feb 13. J Virol. 2008. PMID: 18272586 Free PMC article.
-
Structure and functional relevance of a transcription-regulating sequence involved in coronavirus discontinuous RNA synthesis.J Virol. 2011 May;85(10):4963-73. doi: 10.1128/JVI.02317-10. Epub 2011 Mar 9. J Virol. 2011. PMID: 21389138 Free PMC article.
-
Coronaviruses use discontinuous extension for synthesis of subgenome-length negative strands.Adv Exp Med Biol. 1995;380:499-506. doi: 10.1007/978-1-4615-1899-0_79. Adv Exp Med Biol. 1995. PMID: 8830530 Review.
-
A new model for coronavirus transcription.Adv Exp Med Biol. 1998;440:215-9. doi: 10.1007/978-1-4615-5331-1_26. Adv Exp Med Biol. 1998. PMID: 9782283 Review.
Cited by
-
The SARS-CoV-2 subgenome landscape and its novel regulatory features.Mol Cell. 2021 May 20;81(10):2135-2147.e5. doi: 10.1016/j.molcel.2021.02.036. Epub 2021 Mar 3. Mol Cell. 2021. PMID: 33713597 Free PMC article.
-
Functional transcriptional regulatory sequence (TRS) RNA binding and helix destabilizing determinants of murine hepatitis virus (MHV) nucleocapsid (N) protein.J Biol Chem. 2012 Mar 2;287(10):7063-73. doi: 10.1074/jbc.M111.287763. Epub 2012 Jan 12. J Biol Chem. 2012. PMID: 22241479 Free PMC article.
-
Recent Progress in Torovirus Molecular Biology.Viruses. 2021 Mar 8;13(3):435. doi: 10.3390/v13030435. Viruses. 2021. PMID: 33800523 Free PMC article. Review.
-
Classification, replication, and transcription of Nidovirales.Front Microbiol. 2024 Jan 24;14:1291761. doi: 10.3389/fmicb.2023.1291761. eCollection 2023. Front Microbiol. 2024. PMID: 38328580 Free PMC article. Review.
-
Identification and subcellular localization of porcine deltacoronavirus accessory protein NS6.Virology. 2016 Dec;499:170-177. doi: 10.1016/j.virol.2016.09.015. Epub 2016 Sep 20. Virology. 2016. PMID: 27661736 Free PMC article.
References
-
- Callebaut, P., I. Correa, M. Pensaert, G. Jiménez, and L. Enjuanes. 1988. Antigenic differentiation between transmissible gastroenteritis virus of swine and a related porcine respiratory coronavirus. J. Gen. Virol. 69:1725-1730. - PubMed
-
- Choi, K. S., P.-Y. Huang, and M. C. C. Lai. 2002. Polypyrimidine-tract-binding protein affects transcription but not translation of mouse hepatitis virus RNA. Virology 303:58-68. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

