Distinct mechanisms determine transposon inheritance and methylation via small interfering RNA and histone modification
- PMID: 14691539
- PMCID: PMC300680
- DOI: 10.1371/journal.pbio.0000067
Distinct mechanisms determine transposon inheritance and methylation via small interfering RNA and histone modification
Abstract
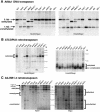
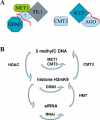
Heritable, but reversible, changes in transposable element activity were first observed in maize by Barbara McClintock in the 1950s. More recently, transposon silencing has been associated with DNA methylation, histone H3 lysine-9 methylation (H3mK9), and RNA interference (RNAi). Using a genetic approach, we have investigated the role of these modifications in the epigenetic regulation and inheritance of six Arabidopsis transposons. Silencing of most of the transposons is relieved in DNA methyltransferase (met1), chromatin remodeling ATPase (ddm1), and histone modification (sil1) mutants. In contrast, only a small subset of the transposons require the H3mK9 methyltransferase KRYPTONITE, the RNAi gene ARGONAUTE1, and the CXG methyltransferase CHROMOMETHYLASE3. In crosses to wild-type plants, epigenetic inheritance of active transposons varied from mutant to mutant, indicating these genes differ in their ability to silence transposons. According to their pattern of transposon regulation, the mutants can be divided into two groups, which suggests that there are distinct, but interacting, complexes or pathways involved in transposon silencing. Furthermore, different transposons tend to be susceptible to different forms of epigenetic regulation.
Conflict of interest statement
The authors have declared that no conflicts of interest exist.
Figures



Similar articles
-
RNAi-independent de novo DNA methylation revealed in Arabidopsis mutants of chromatin remodeling gene DDM1.Plant J. 2012 Jun;70(5):750-8. doi: 10.1111/j.1365-313X.2012.04911.x. Epub 2012 Mar 6. Plant J. 2012. PMID: 22269081
-
Dependence of heterochromatic histone H3 methylation patterns on the Arabidopsis gene DDM1.Science. 2002 Sep 13;297(5588):1871-3. doi: 10.1126/science.1074950. Epub 2002 Jun 20. Science. 2002. PMID: 12077425
-
Chromatin and siRNA pathways cooperate to maintain DNA methylation of small transposable elements in Arabidopsis.Genome Biol. 2005;6(11):R90. doi: 10.1186/gb-2005-6-11-r90. Epub 2005 Oct 19. Genome Biol. 2005. PMID: 16277745 Free PMC article.
-
Silencing of transposons in plant genomes: kick them when they're down.Genome Biol. 2004;5(12):249. doi: 10.1186/gb-2004-5-12-249. Epub 2004 Nov 16. Genome Biol. 2004. PMID: 15575975 Free PMC article. Review.
-
Small RNAs and transposon silencing in plants.Dev Growth Differ. 2012 Jan;54(1):100-7. doi: 10.1111/j.1440-169X.2011.01309.x. Epub 2011 Dec 12. Dev Growth Differ. 2012. PMID: 22150226 Review.
Cited by
-
Male germline control of transposable elements.Biol Reprod. 2012 May 31;86(5):162, 1-14. doi: 10.1095/biolreprod.111.095463. Print 2012 May. Biol Reprod. 2012. PMID: 22357546 Free PMC article. Review.
-
Epigenetic mechanisms of genomic imprinting: common themes in the regulation of imprinted regions in mammals, plants, and insects.Genet Res Int. 2012;2012:585024. doi: 10.1155/2012/585024. Epub 2012 Feb 15. Genet Res Int. 2012. PMID: 22567394 Free PMC article.
-
Induction of gene silencing by hairpin RNA expression in Tetrahymena thermophila reveals a second small RNA pathway.Mol Cell Biol. 2006 Dec;26(23):8731-42. doi: 10.1128/MCB.01430-06. Epub 2006 Sep 25. Mol Cell Biol. 2006. PMID: 17000759 Free PMC article.
-
Retrotransposons: How the continuous evolutionary front shapes plant genomes for response to heat stress.Front Plant Sci. 2022 Dec 9;13:1064847. doi: 10.3389/fpls.2022.1064847. eCollection 2022. Front Plant Sci. 2022. PMID: 36570931 Free PMC article. Review.
-
Telomeric trans-silencing in Drosophila melanogaster: tissue specificity, development and functional interactions between non-homologous telomeres.PLoS One. 2008 Sep 22;3(9):e3249. doi: 10.1371/journal.pone.0003249. PLoS One. 2008. PMID: 18813361 Free PMC article.
References
-
- Aravin AA, Naumova NM, Tulin AV, Vagin VV, Rozovsky YM, et al. Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germline. Curr Biol. 2001;11:1017–1027. - PubMed
-
- Banks JA, Masson P, Federoff N. Molecular mechanisms in the developmental regulation of the maize suppressor–mutator transposable element. Genes Dev. 1988;2:1364–1380. - PubMed
-
- Brzeski J, Jerzmanowski A. Deficient in DNA methylation 1 (DDM1) defines a novel family of chromatin-remodeling factors. J Biol Chem. 2003;278:823–828. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

