Argonaute protein in the early divergent eukaryote Trypanosoma brucei: control of small interfering RNA accumulation and retroposon transcript abundance
- PMID: 14673174
- PMCID: PMC303348
- DOI: 10.1128/MCB.24.1.420-427.2004
Argonaute protein in the early divergent eukaryote Trypanosoma brucei: control of small interfering RNA accumulation and retroposon transcript abundance
Abstract
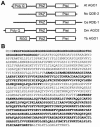
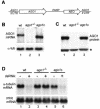
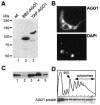
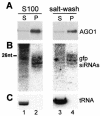
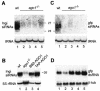
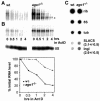
Members of the Argonaute protein family have been linked through a combination of genetic and biochemical studies to RNA interference (RNAi) and related phenomena. Here, we describe the characterization of the first Argonaute protein (AGO1) in Trypanosoma brucei, the earliest divergent eukaryote where RNAi has been described so far. AGO1 is predominantly cytoplasmic and is found in a ribonucleoprotein particle with small interfering RNAs (siRNAs), and this particle is present in a soluble form, as well as associated with polyribosomes. A genetic knockout of AGO1 leads to a loss of RNAi, and concomitantly, endogenous retroposon-derived siRNAs as well as siRNAs derived from transgenic double-stranded RNA are reduced to almost undetectable levels. Furthermore, AGO1 deficiency leads to an increase in retroposon transcript abundance via mechanisms operating at the transcriptional level and at the RNA stability level. Our results suggest that AGO1 function is required for production and/or stabilization of siRNAs and provide the first evidence for an Argonaute protein being involved in the regulation of retroposon transcript levels.
Figures

Similar articles
-
RNA interference in Trypanosoma brucei: cloning of small interfering RNAs provides evidence for retroposon-derived 24-26-nucleotide RNAs.RNA. 2001 Nov;7(11):1522-30. RNA. 2001. PMID: 11720282 Free PMC article.
-
Small interfering RNA-producing loci in the ancient parasitic eukaryote Trypanosoma brucei.BMC Genomics. 2012 Aug 27;13:427. doi: 10.1186/1471-2164-13-427. BMC Genomics. 2012. PMID: 22925482 Free PMC article.
-
RNA interference in Trypanosoma brucei: role of the n-terminal RGG domain and the polyribosome association of argonaute.J Biol Chem. 2009 Dec 25;284(52):36511-36520. doi: 10.1074/jbc.M109.073072. Epub 2009 Oct 30. J Biol Chem. 2009. PMID: 19880512 Free PMC article.
-
RNA interference: advances and questions.Philos Trans R Soc Lond B Biol Sci. 2002 Jan 29;357(1417):65-70. doi: 10.1098/rstb.2001.0952. Philos Trans R Soc Lond B Biol Sci. 2002. PMID: 11839183 Free PMC article. Review.
-
RNA interference in protozoan parasites.Cell Microbiol. 2004 Jun;6(6):509-19. doi: 10.1111/j.1462-5822.2004.00399.x. Cell Microbiol. 2004. PMID: 15104593 Review.
Cited by
-
The structure and repertoire of small interfering RNAs in Leishmania (Viannia) braziliensis reveal diversification in the trypanosomatid RNAi pathway.Mol Microbiol. 2013 Feb;87(3):580-93. doi: 10.1111/mmi.12117. Epub 2012 Dec 26. Mol Microbiol. 2013. PMID: 23217017 Free PMC article.
-
Comparative genomics reveals two novel RNAi factors in Trypanosoma brucei and provides insight into the core machinery.PLoS Pathog. 2012;8(5):e1002678. doi: 10.1371/journal.ppat.1002678. Epub 2012 May 24. PLoS Pathog. 2012. PMID: 22654659 Free PMC article.
-
On the extent and role of the small proteome in the parasitic eukaryote Trypanosoma brucei.BMC Biol. 2014 Feb 19;12:14. doi: 10.1186/1741-7007-12-14. BMC Biol. 2014. PMID: 24552149 Free PMC article.
-
Functionally related transcripts have common RNA motifs for specific RNA-binding proteins in trypanosomes.BMC Mol Biol. 2008 Dec 8;9:107. doi: 10.1186/1471-2199-9-107. BMC Mol Biol. 2008. PMID: 19063746 Free PMC article.
-
Role of the Trypanosoma brucei HEN1 family methyltransferase in small interfering RNA modification.Eukaryot Cell. 2014 Jan;13(1):77-86. doi: 10.1128/EC.00233-13. Epub 2013 Nov 1. Eukaryot Cell. 2014. PMID: 24186950 Free PMC article.
References
-
- Bastin, P., Z. Bagherzadeh, K. R. Matthews, and K. Gull. 1996. A novel epitope tag system to study protein targeting and organelle biogenesis in Trypanosoma brucei. Mol. Biochem. Parasitol. 77:235-239. - PubMed
-
- Carmell, M. A., Z. Xuan, M. Q. Zhang, and G. J. Hannon. 2002. The Argonaute family: tentacles that reach into RNAi, developmental control, stem cell maintenance, and tumorigenesis. Genes Dev. 16:2733-2742. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
