The mouse Murr1 gene is imprinted in the adult brain, presumably due to transcriptional interference by the antisense-oriented U2af1-rs1 gene
- PMID: 14673161
- PMCID: PMC303337
- DOI: 10.1128/MCB.24.1.270-279.2004
The mouse Murr1 gene is imprinted in the adult brain, presumably due to transcriptional interference by the antisense-oriented U2af1-rs1 gene
Abstract
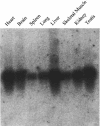
The mouse Murr1 gene contains an imprinted gene, U2af1-rs1, in its first intron. U2af1-rs1 shows paternal allele-specific expression and is transcribed in the direction opposite to that of the Murr1 gene. In contrast to a previous report of biallelic expression of Murr1 in neonatal mice, we have found that the maternal allele is expressed predominantly in the adult brain and also preferentially in other adult tissues. This maternal-predominant expression is not observed in embryonic and neonatal brains. In situ hybridization experiments that used the adult brain indicated that Murr1 gene was maternally expressed in neuronal cells in all regions of the brain. We analyzed the developmental change in the expression levels of both Murr1 and U2af1-rs1 in the brain and liver, and we propose that the maternal-predominant expression of Murr1 results from transcriptional interference of the gene by U2af1-rs1 through the Murr1 promoter region.
Figures


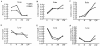

Similar articles
-
Comparative analyses of genomic imprinting and CpG island-methylation in mouse Murr1 and human MURR1 loci revealed a putative imprinting control region in mice.Gene. 2006 Jan 17;366(1):77-86. doi: 10.1016/j.gene.2005.08.020. Epub 2005 Nov 21. Gene. 2006. PMID: 16305817
-
Mouse U2af1-rs1 is a neomorphic imprinted gene.Mol Cell Biol. 1997 Feb;17(2):789-98. doi: 10.1128/MCB.17.2.789. Mol Cell Biol. 1997. PMID: 9001233 Free PMC article.
-
DNA methylation is linked to deacetylation of histone H3, but not H4, on the imprinted genes Snrpn and U2af1-rs1.Mol Cell Biol. 2001 Aug;21(16):5426-36. doi: 10.1128/MCB.21.16.5426-5436.2001. Mol Cell Biol. 2001. PMID: 11463825 Free PMC article.
-
The conflict theory of genomic imprinting: how much can be explained?Curr Top Dev Biol. 1998;40:255-93. doi: 10.1016/s0070-2153(08)60369-5. Curr Top Dev Biol. 1998. PMID: 9673853 Review.
-
Imprinted gene expression in the brain.Neurosci Biobehav Rev. 2005 May;29(3):421-30. doi: 10.1016/j.neubiorev.2004.11.007. Epub 2005 Jan 18. Neurosci Biobehav Rev. 2005. PMID: 15820547 Review.
Cited by
-
The human retinoblastoma gene is imprinted.PLoS Genet. 2009 Dec;5(12):e1000790. doi: 10.1371/journal.pgen.1000790. Epub 2009 Dec 24. PLoS Genet. 2009. PMID: 20041224 Free PMC article.
-
Genomic imprinting in mammals: emerging themes and established theories.PLoS Genet. 2006 Nov 24;2(11):e147. doi: 10.1371/journal.pgen.0020147. PLoS Genet. 2006. PMID: 17121465 Free PMC article. Review.
-
Sex-Dimorphic Behavioral Alterations and Altered Neurogenesis in U12 Intron Splicing-Defective Zrsr1 Mutant Mice.Int J Mol Sci. 2019 Jul 19;20(14):3543. doi: 10.3390/ijms20143543. Int J Mol Sci. 2019. PMID: 31331069 Free PMC article.
-
Genomic imprinting in mouse blastocysts is predominantly associated with H3K27me3.Nat Commun. 2021 Jun 21;12(1):3804. doi: 10.1038/s41467-021-23510-4. Nat Commun. 2021. PMID: 34155196 Free PMC article.
-
Growing oocyte-specific transcription-dependent de novo DNA methylation at the imprinted Zrsr1-DMR.Epigenetics Chromatin. 2018 Jun 6;11(1):28. doi: 10.1186/s13072-018-0200-6. Epigenetics Chromatin. 2018. PMID: 29875017 Free PMC article.
References
-
- Albrecht, U., J. S. Sutcliffe, B. M. Cattanach, C. V. Beechey, D. Armstrong, G. Eichele, and A. L. Beaudet. 1997. Imprinted expression of the murine Angelman syndrome gene, Ube3a, in hippocampal and Purkinje neurons. Nat. Genet. 17:75-78. - PubMed
-
- Avner, P., and E. Heard. 2001. X-chromosome inactivation: counting, choice and initiation. Nat. Rev. Genet. 2:59-67. - PubMed
-
- Barlow, D. P. 1995. Gametic imprinting in mammals. Science 270:1610-1613. - PubMed
-
- Beechey, C. V. 1999. Imprinted genes and regions in mouse and human, p. 303-323. In R. Ohlsson (ed.), Genomic imprinting: an interdisciplinary approach. Results and problems in cell differentiation. Springer-Verlag, Heidelberg, Germany. - PubMed
-
- Cattanach, B. M., and M. Kirk. 1985. Differential activity of maternally and paternally derived chromosome regions in mice. Nature 315:496-498. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
