Determination of the substrate-docking site of protein tyrosine kinase C-terminal Src kinase
- PMID: 14657361
- PMCID: PMC299771
- DOI: 10.1073/pnas.2534493100
Determination of the substrate-docking site of protein tyrosine kinase C-terminal Src kinase
Abstract
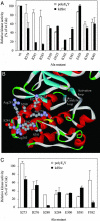
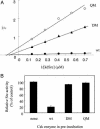
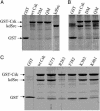
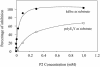
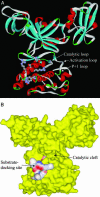
Protein tyrosine kinases (PTK) are key enzymes of mammalian signal transduction. For the fidelity of signal transduction, each PTK phosphorylates only one or a few proteins on specific Tyr residues. Substrate specificity is thought to be mediated by PTK-substrate docking interactions and recognition of the phosphorylation site sequence by the kinase active site. However, a substrate-docking site has not been determined on any PTK. C-terminal Src kinase (Csk) is a PTK that specifically phosphorylates Src family kinases on a C-terminal Tyr. In this study, by sequence alignment and site-specific mutagenesis, we located a substrate-docking site on Csk. Mutations in the docking site disabled Csk to phosphorylate, regulate, and complex with Src but only moderately affected its general kinase activity. A peptide mimicking the docking site potently inhibited (IC50 = 21 microM) Csk phosphorylation of Src but only moderately inhibited (IC50 = 422 microM) its general kinase activity. Determination of the substrate-docking site provides the structural basis of substrate specificity in Csk and a model for understanding substrate specificity in other PTKs.
Figures

Similar articles
-
Dissection of the catalytic and regulatory structure-function relationships of Csk protein tyrosine kinase.Front Cell Dev Biol. 2023 Mar 1;11:1148352. doi: 10.3389/fcell.2023.1148352. eCollection 2023. Front Cell Dev Biol. 2023. PMID: 36936693 Free PMC article. Review.
-
Docking-based substrate recognition by the catalytic domain of a protein tyrosine kinase, C-terminal Src kinase (Csk).J Biol Chem. 2006 Mar 24;281(12):8183-9. doi: 10.1074/jbc.M508120200. Epub 2006 Jan 26. J Biol Chem. 2006. PMID: 16439366
-
Detection of a physical and functional interaction between Csk and Lck which involves the SH2 domain of Csk and is mediated by autophosphorylation of Lck on tyrosine 394.J Biol Chem. 1996 Mar 29;271(13):7465-72. doi: 10.1074/jbc.271.13.7465. J Biol Chem. 1996. PMID: 8631775
-
Structural basis for the recognition of c-Src by its inactivator Csk.Cell. 2008 Jul 11;134(1):124-34. doi: 10.1016/j.cell.2008.05.051. Cell. 2008. PMID: 18614016 Free PMC article.
-
Recognition and specificity in protein tyrosine kinase-mediated signalling.Trends Biochem Sci. 1995 Nov;20(11):470-5. doi: 10.1016/s0968-0004(00)89103-3. Trends Biochem Sci. 1995. PMID: 8578591 Review.
Cited by
-
Dissection of the catalytic and regulatory structure-function relationships of Csk protein tyrosine kinase.Front Cell Dev Biol. 2023 Mar 1;11:1148352. doi: 10.3389/fcell.2023.1148352. eCollection 2023. Front Cell Dev Biol. 2023. PMID: 36936693 Free PMC article. Review.
-
Characterization of the interactions between the active site of a protein tyrosine kinase and a divalent metal activator.BMC Biochem. 2005 Nov 23;6:25. doi: 10.1186/1471-2091-6-25. BMC Biochem. 2005. PMID: 16305747 Free PMC article.
-
Identification of N-terminal lobe motifs that determine the kinase activity of the catalytic domains and regulatory strategies of Src and Csk protein tyrosine kinases.J Mol Biol. 2009 Mar 6;386(4):1066-77. doi: 10.1016/j.jmb.2009.01.012. J Mol Biol. 2009. PMID: 19244618 Free PMC article.
-
Metalloproteinase/Presenilin1 processing of ephrinB regulates EphB-induced Src phosphorylation and signaling.EMBO J. 2006 Mar 22;25(6):1242-52. doi: 10.1038/sj.emboj.7601031. Epub 2006 Mar 2. EMBO J. 2006. PMID: 16511561 Free PMC article.
-
Signaling properties of a non-metazoan Src kinase and the evolutionary history of Src negative regulation.J Biol Chem. 2008 May 30;283(22):15491-501. doi: 10.1074/jbc.M800002200. Epub 2008 Apr 4. J Biol Chem. 2008. PMID: 18390552 Free PMC article.
References
-
- Manning, G., Whyte, D. B., Martinez, R., Hunter, T. & Sudarsanam, S. (2002) Science 298, 1912–1934. - PubMed
-
- Hunter, T. (1995) Cell 80, 225–236. - PubMed
-
- Blume-Jensen, P. & Hunter, T. (2001) Nature 411, 355–365. - PubMed
-
- Capdeville, R., Buchdunger, E., Zimmermann, J. & Matter, A. (2002) Nat. Rev. Drug Discov. 1, 493–502. - PubMed
-
- Hubbard, S. R. & Till, J. H. (2000) Annu. Rev. Biochem. 69, 373–398. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

