Visualizing telomere dynamics in living mammalian cells using PNA probes
- PMID: 14657034
- PMCID: PMC291828
- DOI: 10.1093/emboj/cdg633
Visualizing telomere dynamics in living mammalian cells using PNA probes
Abstract
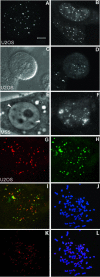
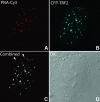
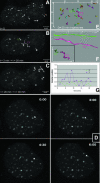
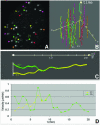
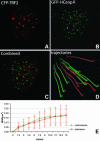
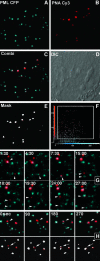
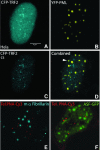
Chromosome ends are protected from degradation by the presence of the highly repetitive hexanucleotide sequence of TTAGGG and associated proteins. These so-called telomeric complexes are suggested to play an important role in establishing a functional nuclear chromatin organization. Using peptide nucleic acid (PNA) probes, we studied the dynamic behavior of telomeric DNA repeats in living human osteosarcoma U2OS cells. A fluorescent cy3-labeled PNA probe was introduced in living cells by glass bead loading and was shown to specifically associate with telomeric DNA shortly afterwards. Telomere dynamics were imaged for several hours using digital fluorescence microscopy. While the majority of telomeres revealed constrained diffusive movement, individual telomeres in a human cell nucleus showed significant directional movements. Also, a subfraction of telomeres were shown to associate and dissociate, suggesting that in vivo telomere clusters are not stable but dynamic structures. Furthermore, telomeres were shown to associate with promyelocytic leukemia (PML) bodies in a dynamic manner.
Figures


Similar articles
-
A negative regulator of telomere-length protein trf1 is associated with interstitial (TTAGGG)n blocks in immortal Chinese hamster ovary cells.Biochem Biophys Res Commun. 2001 Jan 19;280(2):471-5. doi: 10.1006/bbrc.2000.4143. Biochem Biophys Res Commun. 2001. PMID: 11162541
-
Telomere-telomere interactions and candidate telomere binding protein(s) in mammalian sperm cells.Exp Cell Res. 1997 Apr 10;232(1):29-41. doi: 10.1006/excr.1997.3482. Exp Cell Res. 1997. PMID: 9141618
-
Comet-FISH using peptide nucleic acid probes detects telomeric repeats in DNA damaged by bleomycin and mitomycin C proportional to general DNA damage.Mutagenesis. 2004 Sep;19(5):403-8. doi: 10.1093/mutage/geh049. Mutagenesis. 2004. PMID: 15388814
-
Measurement of telomere length using PNA probe by cytometry.Methods Cell Biol. 2011;103:189-202. doi: 10.1016/B978-0-12-385493-3.00008-5. Methods Cell Biol. 2011. PMID: 21722804 Review.
-
Budding yeast with human telomeres: a puzzling structure.Biochimie. 2008 Jan;90(1):108-15. doi: 10.1016/j.biochi.2007.09.009. Epub 2007 Sep 22. Biochimie. 2008. PMID: 17954006 Review.
Cited by
-
Mobility of multi-subunit complexes in the nucleus: accessibility and dynamics of chromatin subcompartments.Histochem Cell Biol. 2005 Mar;123(3):217-28. doi: 10.1007/s00418-005-0752-y. Epub 2005 Apr 14. Histochem Cell Biol. 2005. PMID: 15830242 Review.
-
Probing local chromatin dynamics by tracking telomeres.Biophys J. 2022 Jul 19;121(14):2684-2692. doi: 10.1016/j.bpj.2022.06.020. Epub 2022 Jun 22. Biophys J. 2022. PMID: 35733342 Free PMC article.
-
Uniform Contraction-Expansion Description of Relative Centromere and Telomere Motion.Biophys J. 2015 Oct 6;109(7):1454-62. doi: 10.1016/j.bpj.2015.07.031. Biophys J. 2015. PMID: 26445446 Free PMC article.
-
DNA/RNA Fluorescence Imaging by Synthetic Nucleic Acids.Adv Exp Med Biol. 2021;1310:475-493. doi: 10.1007/978-981-33-6064-8_17. Adv Exp Med Biol. 2021. PMID: 33834446
-
Characterization of telomeric repeats in metaphase chromosomes and interphase nuclei of Syrian Hamster Fibroblasts.Mol Cytogenet. 2012 Sep 3;5(1):37. doi: 10.1186/1755-8166-5-37. Mol Cytogenet. 2012. PMID: 22938505 Free PMC article.
References
-
- Azzalin C.M., Nergadze,S.G. and Giulotto,E. (2001) Human interchromosomal telomeric-like repeats: sequence organization and mechanisms of origin. Chromosoma, 110, 75–82. - PubMed
-
- Baur J.A., Zou,Y., Shay,J.W. and Wright,W.E. (2001) Telomere position effect in human cells. Science, 292, 2075–2077. - PubMed
-
- Brown K.E., Baxter,J., Graf,D., Merkenschlager,M. and Fisher,A.G. (1999) Dynamic repositioning of genes in the nucleus of lymphocytes preparing for cell division. Mol. Cell, 3, 207–217. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

