Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus
- PMID: 14647384
- PMCID: PMC7095016
- DOI: 10.1038/nature02145
Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus
Abstract
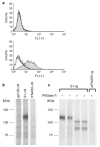
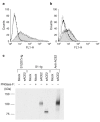
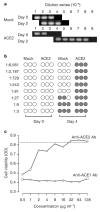
Spike (S) proteins of coronaviruses, including the coronavirus that causes severe acute respiratory syndrome (SARS), associate with cellular receptors to mediate infection of their target cells. Here we identify a metallopeptidase, angiotensin-converting enzyme 2 (ACE2), isolated from SARS coronavirus (SARS-CoV)-permissive Vero E6 cells, that efficiently binds the S1 domain of the SARS-CoV S protein. We found that a soluble form of ACE2, but not of the related enzyme ACE1, blocked association of the S1 domain with Vero E6 cells. 293T cells transfected with ACE2, but not those transfected with human immunodeficiency virus-1 receptors, formed multinucleated syncytia with cells expressing S protein. Furthermore, SARS-CoV replicated efficiently on ACE2-transfected but not mock-transfected 293T cells. Finally, anti-ACE2 but not anti-ACE1 antibody blocked viral replication on Vero E6 cells. Together our data indicate that ACE2 is a functional receptor for SARS-CoV.
Conflict of interest statement
The authors declare that they have no competing financial interests.
Figures

Similar articles
-
Retroviruses pseudotyped with the severe acute respiratory syndrome coronavirus spike protein efficiently infect cells expressing angiotensin-converting enzyme 2.J Virol. 2004 Oct;78(19):10628-35. doi: 10.1128/JVI.78.19.10628-10635.2004. J Virol. 2004. PMID: 15367630 Free PMC article.
-
Differential downregulation of ACE2 by the spike proteins of severe acute respiratory syndrome coronavirus and human coronavirus NL63.J Virol. 2010 Jan;84(2):1198-205. doi: 10.1128/JVI.01248-09. Epub 2009 Oct 28. J Virol. 2010. PMID: 19864379 Free PMC article.
-
Importance of cholesterol-rich membrane microdomains in the interaction of the S protein of SARS-coronavirus with the cellular receptor angiotensin-converting enzyme 2.Virology. 2008 Nov 25;381(2):215-21. doi: 10.1016/j.virol.2008.08.026. Epub 2008 Sep 23. Virology. 2008. PMID: 18814896 Free PMC article.
-
Insights from the association of SARS-CoV S-protein with its receptor, ACE2.Adv Exp Med Biol. 2006;581:209-18. doi: 10.1007/978-0-387-33012-9_36. Adv Exp Med Biol. 2006. PMID: 17037532 Free PMC article. Review. No abstract available.
-
Angiotensin-converting enzyme 2: a functional receptor for SARS coronavirus.Cell Mol Life Sci. 2004 Nov;61(21):2738-43. doi: 10.1007/s00018-004-4242-5. Cell Mol Life Sci. 2004. PMID: 15549175 Free PMC article. Review.
Cited by
-
Gender Differences in Psychological and Behavioral Responses of Infected and Uninfected Health-Care Workers During the Early COVID-19 Outbreak.Front Public Health. 2021 Mar 11;9:638975. doi: 10.3389/fpubh.2021.638975. eCollection 2021. Front Public Health. 2021. PMID: 33777887 Free PMC article.
-
Impact of cytokine storm and systemic inflammation on liver impairment patients infected by SARS-CoV-2: Prospective therapeutic challenges.World J Gastroenterol. 2021 Apr 21;27(15):1531-1552. doi: 10.3748/wjg.v27.i15.1531. World J Gastroenterol. 2021. PMID: 33958841 Free PMC article. Review.
-
Pharmacological approaches to the treatment of COVID-19 patients.J Transl Sci. 2020 Dec;6(6):394. Epub 2020 Apr 17. J Transl Sci. 2020. PMID: 33042589 Free PMC article.
-
ACE2, TMPRSS2, and Furin variants and SARS-CoV-2 infection in Madrid, Spain.J Med Virol. 2021 Feb;93(2):863-869. doi: 10.1002/jmv.26319. Epub 2020 Jul 28. J Med Virol. 2021. PMID: 32691890 Free PMC article.
-
COVID-19 and the Digestive System.Am J Gastroenterol. 2020 Jul;115(7):1003-1006. doi: 10.14309/ajg.0000000000000691. Am J Gastroenterol. 2020. PMID: 32618648 Free PMC article. Review.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

