GATA-4 and GATA-5 transcription factor genes and potential downstream antitumor target genes are epigenetically silenced in colorectal and gastric cancer
- PMID: 14612389
- PMCID: PMC262684
- DOI: 10.1128/MCB.23.23.8429-8439.2003
GATA-4 and GATA-5 transcription factor genes and potential downstream antitumor target genes are epigenetically silenced in colorectal and gastric cancer
Abstract
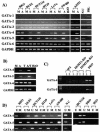
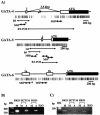
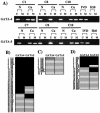
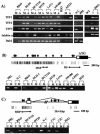
The GATA family of transcription factors participates in gastrointestinal (GI) development. Increases in GATA-4 and -5 expression occur in differentiation and GATA-6 expression in proliferation in embryonic and adult settings. We now show that in colorectal cancer (CRC) and gastric cancer promoter hypermethylation and transcriptional silencing are frequent for GATA-4 and -5 but are never seen for GATA-6. Potential antitumor target genes upregulated by GATA-4 and -5, the trefoil factors, inhibinalpha, and disabled-2 (Dab2) are also silenced, in GI cancers, with associated methylation of the promoters. Drug or genetically induced demethylation simultaneously leads to expression, in CRC cells, of all of the GATA-4, -5, and downstream genes. Expression of exogenous GATA-5 overrides methylation at the downstream promoters to activate the target genes. Selection for silencing of both upstream transcription factors and their target genes in GI cancers could indicate that epigenetic silencing of the involved genes provides a summated contribution to tumor progression.
Figures


Similar articles
-
Hypermethylation of the GATA genes in lung cancer.Clin Cancer Res. 2004 Dec 1;10(23):7917-24. doi: 10.1158/1078-0432.CCR-04-1140. Clin Cancer Res. 2004. PMID: 15585625
-
Hypermethylation of the GATA gene family in esophageal cancer.Int J Cancer. 2006 Nov 1;119(9):2078-83. doi: 10.1002/ijc.22092. Int J Cancer. 2006. PMID: 16823849
-
Involvement of GATA-4/-5 transcription factors in ovarian carcinogenesis.Cancer Lett. 2006 Sep 28;241(2):281-8. doi: 10.1016/j.canlet.2005.10.039. Epub 2005 Dec 7. Cancer Lett. 2006. PMID: 16337738
-
Gene methylation in gastric cancer.Clin Chim Acta. 2013 Sep 23;424:53-65. doi: 10.1016/j.cca.2013.05.002. Epub 2013 May 10. Clin Chim Acta. 2013. PMID: 23669186 Review.
-
The roles of GATA-4, -5 and -6 in vertebrate heart development.Semin Cell Dev Biol. 2005 Feb;16(1):83-94. doi: 10.1016/j.semcdb.2004.10.003. Epub 2004 Dec 15. Semin Cell Dev Biol. 2005. PMID: 15659343 Review.
Cited by
-
Upstream stimulatory factor 1 activates GATA5 expression through an E-box motif.Biochem J. 2012 Aug 15;446(1):89-98. doi: 10.1042/BJ20111942. Biochem J. 2012. PMID: 22625849 Free PMC article.
-
Loss of disabled-2 expression is an early event in esophageal squamous tumorigenesis.World J Gastroenterol. 2006 Oct 7;12(37):6041-5. doi: 10.3748/wjg.v12.i37.6041. World J Gastroenterol. 2006. PMID: 17009406 Free PMC article.
-
Aberrant gene promoter methylation associated with sporadic multiple colorectal cancer.PLoS One. 2010 Jan 19;5(1):e8777. doi: 10.1371/journal.pone.0008777. PLoS One. 2010. PMID: 20098741 Free PMC article.
-
Distinct expression and prognostic values of GATA transcription factor family in human ovarian cancer.J Ovarian Res. 2022 Apr 29;15(1):49. doi: 10.1186/s13048-022-00974-6. J Ovarian Res. 2022. PMID: 35488350 Free PMC article.
-
A four-DNA methylation biomarker is a superior predictor of survival of patients with cutaneous melanoma.Elife. 2019 Jun 6;8:e44310. doi: 10.7554/eLife.44310. Elife. 2019. PMID: 31169496 Free PMC article.
References
-
- Al-Azzeh, E. D., P. Fegert, N. Blin, and P. Gott. 2000. Transcription factor GATA-6 activates expression of gastroprotective trefoil genes TFF1 and TFF2. Biochim. Biophys. Acta 1490:324-332. - PubMed
-
- Azarschab, P., E. Al-Azzeh, W. Kornberger, and P. Gott. 2001. Aspirin promotes TFF2 gene activation in human gastric cancer cell lines. FEBS Lett. 488:206-210. - PubMed
-
- Bai, Y., Y. Akiyama, H. Nagasaki, O. K. Yagi, Y. Kikuchi, N. Saito, K. Takeshita, T. Iwai, and Y. Yuasa. 2000. Distinct expression of CDX2 and GATA4/5, development-related genes, in human gastric cancer cell lines. Mol. Carcinog. 28:184-188. - PubMed
-
- Bird, A. 2002. DNA methylation patterns and epigenetic memory. Genes Dev. 16:6-21. - PubMed
-
- Boudreau, F., E. H. Rings, H. M. van Wering, R. K. Kim, G. P. Swain, S. D. Krasinski, J. Moffett, R. J. Grand, E. R. Suh, and P. G. Traber. 2002. Hepatocyte nuclear factor-1α, GATA-4, and caudal related homeodomain protein Cdx2 interact functionally to modulate intestinal gene transcription. Implication for the developmental regulation of the sucrase-isomaltase gene. J. Biol. Chem. 277:31909-31917. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
