Whole-genome screening indicates a possible burst of formation of processed pseudogenes and Alu repeats by particular L1 subfamilies in ancestral primates
- PMID: 14611660
- PMCID: PMC329124
- DOI: 10.1186/gb-2003-4-11-r74
Whole-genome screening indicates a possible burst of formation of processed pseudogenes and Alu repeats by particular L1 subfamilies in ancestral primates
Abstract
Background: Abundant pseudogenes are a feature of mammalian genomes. Processed pseudogenes (PPs) are reverse transcribed from mRNAs. Recent molecular biological studies show that mammalian long interspersed element 1 (L1)-encoded proteins may have been involved in PP reverse transcription. Here, we present the first comprehensive analysis of human PPs using all known human genes as queries.
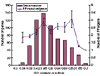
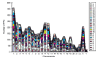
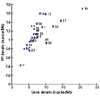
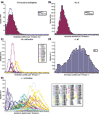
Results: The human genome was queried and 3,664 candidate PPs were identified. The most abundant were copies of genes encoding keratin 18, glyceraldehyde-3-phosphate dehydrogenase and ribosomal protein L21. A simple method was developed to estimate the level of nucleotide substitutions (and therefore the age) of PPs. A Poisson-like age distribution was obtained with a mean age close to that of the Alu repeats, the predominant human short interspersed elements. These data suggest a nearly simultaneous burst of PP and Alu formation in the genomes of ancestral primates. The peak period of amplification of these two distinct retrotransposons was estimated to be 40-50 million years ago. Concordant amplification of certain L1 subfamilies with PPs and Alus was observed.
Conclusions: We suggest that a burst of formation of PPs and Alus occurred in the genome of ancestral primates. One possible mechanism is that proteins encoded by members of particular L1 subfamilies acquired an enhanced ability to recognize cytosolic RNAs in trans.
Figures

Similar articles
-
Novel Role of 3'UTR-Embedded Alu Elements as Facilitators of Processed Pseudogene Genesis and Host Gene Capture by Viral Genomes.PLoS One. 2016 Dec 29;11(12):e0169196. doi: 10.1371/journal.pone.0169196. eCollection 2016. PLoS One. 2016. PMID: 28033411 Free PMC article.
-
L1 elements, processed pseudogenes and retrogenes in mammalian genomes.IUBMB Life. 2006 Dec;58(12):677-85. doi: 10.1080/15216540601034856. IUBMB Life. 2006. PMID: 17424906 Review.
-
Comparative analysis of processed pseudogenes in the mouse and human genomes.Trends Genet. 2004 Feb;20(2):62-7. doi: 10.1016/j.tig.2003.12.005. Trends Genet. 2004. PMID: 14746985 Review.
-
The (r)evolution of SINE versus LINE distributions in primate genomes: sex chromosomes are important.Genome Res. 2010 May;20(5):600-13. doi: 10.1101/gr.099044.109. Epub 2010 Mar 10. Genome Res. 2010. PMID: 20219940 Free PMC article.
-
Recognition of 3'-end L1, Alu, processed pseudogenes, and mRNA stem-loops in the human genome using sequence-based and structure-based machine-learning models.Sci Rep. 2019 May 10;9(1):7211. doi: 10.1038/s41598-019-43403-3. Sci Rep. 2019. PMID: 31076573 Free PMC article.
Cited by
-
The DNA Habitat and its RNA Inhabitants: At the Dawn of RNA Sociology.Genomics Insights. 2013 Mar 4;6:1-12. doi: 10.4137/GEI.S11490. eCollection 2013. Genomics Insights. 2013. PMID: 26217106 Free PMC article.
-
A tale of an A-tail: The lifeline of a SINE.Mob Genet Elements. 2012 Nov 1;2(6):282-286. doi: 10.4161/mge.23204. Mob Genet Elements. 2012. PMID: 23481375 Free PMC article. No abstract available.
-
Independent mammalian genome contractions following the KT boundary.Genome Biol Evol. 2009 May 5;1:2-12. doi: 10.1093/gbe/evp007. Genome Biol Evol. 2009. PMID: 20333172 Free PMC article.
-
Retrotransposons evolution and impact on lncRNA and protein coding genes in pigs.Mob DNA. 2019 May 6;10:19. doi: 10.1186/s13100-019-0161-8. eCollection 2019. Mob DNA. 2019. PMID: 31080521 Free PMC article.
-
Analysis of SINE Families B2, Dip, and Ves with Special Reference to Polyadenylation Signals and Transcription Terminators.Int J Mol Sci. 2021 Sep 13;22(18):9897. doi: 10.3390/ijms22189897. Int J Mol Sci. 2021. PMID: 34576060 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

