Tor pathway regulates Rrn3p-dependent recruitment of yeast RNA polymerase I to the promoter but does not participate in alteration of the number of active genes
- PMID: 14595104
- PMCID: PMC329406
- DOI: 10.1091/mbc.e03-08-0594
Tor pathway regulates Rrn3p-dependent recruitment of yeast RNA polymerase I to the promoter but does not participate in alteration of the number of active genes
Abstract
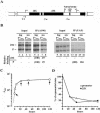
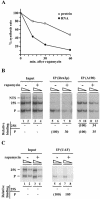
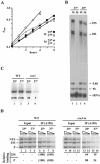
Yeast cells entering into stationary phase decrease rRNA synthesis rate by decreasing both the number of active genes and the transcription rate of individual active genes. Using chromatin immunoprecipitation assays, we found that the association of RNA polymerase I with the promoter and the coding region of rDNA is decreased in stationary phase, but association of transcription factor UAF with the promoter is unchanged. Similar changes were also observed when growing cells were treated with rapamycin, which is known to inhibit the Tor signaling system. Rapamycin treatment also caused a decrease in the amount of Rrn3p-polymerase I complex, similar to stationary phase. Because recruitment of Pol I to the rDNA promoter is Rrn3p-dependent as shown in this work, these data suggest that the decrease in the transcription rate of individual active genes in stationary phase is achieved by the Tor signaling system acting at the Rrn3p-dependent polymerase recruitment step. Miller chromatin spreads of cells treated with rapamycin and cells in post-log phase confirm this conclusion and demonstrate that the Tor system does not participate in alteration of the number of active genes observed for cells entering into stationary phase.
Figures
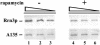


Similar articles
-
The composition of the RNA polymerase I transcription machinery switches from initiation to elongation mode.FEBS Lett. 2004 Apr 23;564(1-2):41-6. doi: 10.1016/S0014-5793(04)00311-4. FEBS Lett. 2004. PMID: 15094040
-
TOR-dependent reduction in the expression level of Rrn3p lowers the activity of the yeast RNA Pol I machinery, but does not account for the strong inhibition of rRNA production.Nucleic Acids Res. 2010 Sep;38(16):5315-26. doi: 10.1093/nar/gkq264. Epub 2010 Apr 25. Nucleic Acids Res. 2010. PMID: 20421203 Free PMC article.
-
New model for the yeast RNA polymerase I transcription cycle.Mol Cell Biol. 2001 Aug;21(15):4847-55. doi: 10.1128/MCB.21.15.4847-4855.2001. Mol Cell Biol. 2001. PMID: 11438642 Free PMC article.
-
The fission yeast RPA21 subunit of RNA polymerase I: an evolutionarily conserved subunit interacting with ribosomal DNA (rDNA) transcription factor Rrn3p for recruitment to rDNA promoter.Genes Genet Syst. 2002 Jun;77(3):147-57. doi: 10.1266/ggs.77.147. Genes Genet Syst. 2002. PMID: 12207036
-
A specialized form of RNA polymerase I, essential for initiation and growth-dependent regulation of rRNA synthesis, is disrupted during transcription.EMBO J. 1998 Jul 1;17(13):3692-703. doi: 10.1093/emboj/17.13.3692. EMBO J. 1998. PMID: 9649439 Free PMC article.
Cited by
-
Sch9 partially mediates TORC1 signaling to control ribosomal RNA synthesis.Cell Cycle. 2009 Dec 15;8(24):4085-90. doi: 10.4161/cc.8.24.10170. Epub 2009 Dec 25. Cell Cycle. 2009. PMID: 19823048 Free PMC article.
-
Transcriptional regulation in yeast during diauxic shift and stationary phase.OMICS. 2010 Dec;14(6):629-38. doi: 10.1089/omi.2010.0069. Epub 2010 Sep 23. OMICS. 2010. PMID: 20863251 Free PMC article. Review.
-
mTOR binds to the promoters of RNA polymerase I- and III-transcribed genes.Cell Cycle. 2010 Mar 1;9(5):953-7. doi: 10.4161/cc.9.5.10876. Epub 2010 Mar 7. Cell Cycle. 2010. PMID: 20038818 Free PMC article.
-
Genome architecture and stability in the Saccharomyces cerevisiae knockout collection.Nature. 2019 Sep;573(7774):416-420. doi: 10.1038/s41586-019-1549-9. Epub 2019 Sep 11. Nature. 2019. PMID: 31511699 Free PMC article.
-
Histones are required for transcription of yeast rRNA genes by RNA polymerase I.Proc Natl Acad Sci U S A. 2005 Jul 19;102(29):10129-34. doi: 10.1073/pnas.0504563102. Epub 2005 Jul 7. Proc Natl Acad Sci U S A. 2005. PMID: 16002464 Free PMC article.
References
-
- Broach, J.R. (1991). RAS genes in Saccharomyces cerevisiae: signal transduction in search of a pathway. Trends Genet. 7, 28-33. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

