DNA basepair step deformability inferred from molecular dynamics simulations
- PMID: 14581192
- PMCID: PMC1303568
- DOI: 10.1016/S0006-3495(03)74710-9
DNA basepair step deformability inferred from molecular dynamics simulations
Abstract
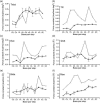
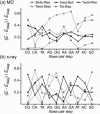
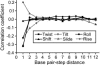
The sequence-dependent DNA deformability at the basepair step level was investigated using large-scale atomic resolution molecular dynamics simulation of two 18-bp DNA oligomers: d(GCCTATAAACGCCTATAA) and d(CTAGGTGGATGACTCATT). From an analysis of the structural fluctuations, the harmonic potential energy functions for all 10 unique steps with respect to the six step parameters have been evaluated. In the case of roll, three distinct groups of steps have been identified: the flexible pyrimidine-purine (YR) steps, intermediate purine-purine (RR), and stiff purine-pyrimidine (RY). The YR steps appear to be the most flexible in tilt and partially in twist. Increasing stiffness from YR through RR to RY was observed for rise, whereas shift and slide lack simple trends. A proposed measure of the relative importance of couplings identifies the slide-rise, twist-roll, and twist-slide couplings to play a major role. The force constants obtained are of similar magnitudes to those based on a crystallographic ensemble. However, the current data have a less complicated and less pronounced sequence dependence. A correlation analysis reveals concerted motions of neighboring steps and thus exposes limitations in the dinucleotide model. The comparison of DNA deformability from this and other studies with recent quantum-chemical stacking energy calculations suggests poor correlation between the stacking and flexibility.
Figures
Similar articles
-
Sequence-dependent DNA structure: the role of the sugar-phosphate backbone.J Mol Biol. 1998 Jul 17;280(3):407-20. doi: 10.1006/jmbi.1998.1865. J Mol Biol. 1998. PMID: 9665845
-
Sequence-dependent dynamics of duplex DNA: the applicability of a dinucleotide model.Biophys J. 2002 Dec;83(6):3446-59. doi: 10.1016/S0006-3495(02)75344-7. Biophys J. 2002. PMID: 12496111 Free PMC article.
-
Sequence-dependent DNA structure: dinucleotide conformational maps.J Mol Biol. 2000 Jan 7;295(1):71-83. doi: 10.1006/jmbi.1999.3236. J Mol Biol. 2000. PMID: 10623509
-
DNA sequence-dependent deformability--insights from computer simulations.Biopolymers. 2004 Feb 15;73(3):327-39. doi: 10.1002/bip.10542. Biopolymers. 2004. PMID: 14755569 Review.
-
Molecular dynamics simulations of DNA curvature and flexibility: helix phasing and premelting.Biopolymers. 2004 Feb 15;73(3):380-403. doi: 10.1002/bip.20019. Biopolymers. 2004. PMID: 14755574 Review.
Cited by
-
Toward a consensus view of duplex RNA flexibility.Biophys J. 2010 Sep 22;99(6):1876-85. doi: 10.1016/j.bpj.2010.06.061. Biophys J. 2010. PMID: 20858433 Free PMC article.
-
DNA methylation of the promoter region at the CREB1 binding site is a mechanism for the epigenetic regulation of brain-specific PKMζ.Biochim Biophys Acta Gene Regul Mech. 2023 Mar;1866(1):194909. doi: 10.1016/j.bbagrm.2023.194909. Epub 2023 Jan 20. Biochim Biophys Acta Gene Regul Mech. 2023. PMID: 36682583 Free PMC article.
-
Sequence-dependent twist-stretch coupling in DNA.Biophys J. 2007 Feb 15;92(4):L30-2. doi: 10.1529/biophysj.106.099572. Epub 2006 Dec 1. Biophys J. 2007. PMID: 17142263 Free PMC article.
-
Intrinsic flexibility of B-DNA: the experimental TRX scale.Nucleic Acids Res. 2010 Jan;38(3):1034-47. doi: 10.1093/nar/gkp962. Epub 2009 Nov 17. Nucleic Acids Res. 2010. PMID: 19920127 Free PMC article.
-
Twist-bend coupling and the statistical mechanics of the twistable wormlike-chain model of DNA: Perturbation theory and beyond.Phys Rev E. 2019 Mar;99(3-1):032414. doi: 10.1103/PhysRevE.99.032414. Phys Rev E. 2019. PMID: 30999490 Free PMC article.
References
-
- Anselmi, C., P. D. Santis, R. Paparcone, M. Savino, and A. Scipioni. 2002. From the sequence to the superstructural properties of DNAs. Biophys. Chem. 95:23–47. - PubMed
-
- Arnott, S., R. Chandrasekaran, D. L. Birdsall, A. G. W. Leslie, and R. L. Ratliff. 1980. Left-handed DNA helices. Nature. 283:743–745. - PubMed
-
- Banavali, N. K., and J. A. D. MacKerell. 2002. Free energy and structural pathways of base flipping in a DNA GCGC containing sequence. J. Mol. Biol. 319:141–160. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

