Cross-repressive interaction of the Olig2 and Nkx2.2 transcription factors in developing neural tube associated with formation of a specific physical complex
- PMID: 14573534
- PMCID: PMC6740479
- DOI: 10.1523/JNEUROSCI.23-29-09547.2003
Cross-repressive interaction of the Olig2 and Nkx2.2 transcription factors in developing neural tube associated with formation of a specific physical complex
Abstract
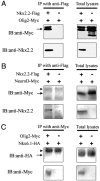
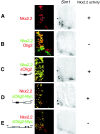
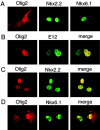
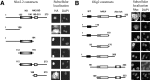
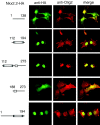
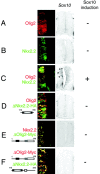
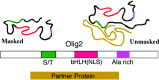
In developing neural tube, the basic helix-loop-helix (bHLH) transcription factor Olig2 interacts with the homeodomain transcription factor Nkx2.2 at two distinct stages. During neuronogenesis, a cross-repressive interaction appears to establish the precise boundary between the p3 and pMN domains. At later times, a cooperative interaction is noted because Nkx2.2 promotes maturation of oligodendrocyte progenitor cells specified by expression of Olig2. We show here that the Olig2 protein can form a physical complex with Nkx2.2 protein in mammalian cells and yeast two-hybrid trap assay. This interaction is specific because Olig2 does not bind to a biologically irrelevant homeodomain transcription factor (Nkx6.1), and Nkx2.2 does not interact with a biologically irrelevant bHLH protein (NeuroD). Deletion mapping analysis suggests that formation of an Olig2-Nkx2.2 physical complex is insufficient for the induction of oligodendrocyte progenitors in developing spine; however, the protein-protein interaction observed might be important for the cross-repressive interaction between Olig2 and Nkx2.2 that helps to establish the pMN-p3 boundary in the developing spinal cord.
Figures


Similar articles
-
The bHLH transcription factor Olig2 promotes oligodendrocyte differentiation in collaboration with Nkx2.2.Neuron. 2001 Sep 13;31(5):791-807. doi: 10.1016/s0896-6273(01)00414-7. Neuron. 2001. PMID: 11567617
-
Mir-17-3p controls spinal neural progenitor patterning by regulating Olig2/Irx3 cross-repressive loop.Neuron. 2011 Feb 24;69(4):721-35. doi: 10.1016/j.neuron.2011.01.014. Neuron. 2011. PMID: 21338882 Free PMC article.
-
Olig bHLH proteins interact with homeodomain proteins to regulate cell fate acquisition in progenitors of the ventral neural tube.Curr Biol. 2001 Sep 18;11(18):1413-20. doi: 10.1016/s0960-9822(01)00441-9. Curr Biol. 2001. PMID: 11566099
-
Induction of oligodendrocyte differentiation by Olig2 and Sox10: evidence for reciprocal interactions and dosage-dependent mechanisms.Dev Biol. 2007 Feb 15;302(2):683-93. doi: 10.1016/j.ydbio.2006.10.007. Epub 2006 Oct 10. Dev Biol. 2007. PMID: 17098222
-
Novel Homeodomain Transcription Factor Nkx2.2 in the Brain Tumor Development.Curr Cancer Drug Targets. 2020;20(5):335-340. doi: 10.2174/1568009618666180102111539. Curr Cancer Drug Targets. 2020. PMID: 29295693 Review.
Cited by
-
Stage-specific deletion of Olig2 conveys opposing functions on differentiation and maturation of oligodendrocytes.J Neurosci. 2013 May 8;33(19):8454-62. doi: 10.1523/JNEUROSCI.2453-12.2013. J Neurosci. 2013. PMID: 23658182 Free PMC article.
-
Combinatorial profiles of oligodendrocyte-selective classes of transcriptional regulators differentially modulate myelin basic protein gene expression.J Neurosci. 2005 Sep 7;25(36):8311-21. doi: 10.1523/JNEUROSCI.1850-05.2005. J Neurosci. 2005. PMID: 16148239 Free PMC article.
-
Molecular Control of Oligodendrocyte Development.Trends Neurosci. 2019 Apr;42(4):263-277. doi: 10.1016/j.tins.2019.01.002. Epub 2019 Feb 12. Trends Neurosci. 2019. PMID: 30770136 Free PMC article. Review.
-
Phosphorylation regulates OLIG2 cofactor choice and the motor neuron-oligodendrocyte fate switch.Neuron. 2011 Mar 10;69(5):918-29. doi: 10.1016/j.neuron.2011.01.030. Neuron. 2011. PMID: 21382552 Free PMC article.
-
Shh and Olig2 sequentially regulate oligodendrocyte differentiation from hiPSCs for the treatment of ischemic stroke.Theranostics. 2022 Mar 28;12(7):3131-3149. doi: 10.7150/thno.69217. eCollection 2022. Theranostics. 2022. PMID: 35547747 Free PMC article.
References
-
- Bertrand N, Castro DS, Guillemot F ( 2002) Proneural genes and the specification of neural cell types. Nat Rev Neurosci 3: 517-530. - PubMed
-
- Briscoe J, Sussel L, Serup P, Hartigan-O'Connor D, Jessell TM, Rubenstein JL, Ericson J ( 1999) Homeobox gene Nkx2.2 and specification of neuronal identity by graded Sonic hedgehog signaling. Nature 398: 622-627. - PubMed
-
- Briscoe J, Pierani A, Jessell TM, Ericson J ( 2000) A homeodomain protein code specifies progenitor cell identity and neuronal fate in the ventral neural tube. Cell 101: 435-445. - PubMed
-
- Ericson J, Rashbass P, Schedl A, Brenner-Morton S, Kawakami A, van Heyningen V, Jessell TM, Briscoe J ( 1997) Pax6 controls progenitor cell identity and neuronal fate in response to graded Shh signaling. Cell 90: 169-180. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

