An endogenous retroviral long terminal repeat is the dominant promoter for human beta1,3-galactosyltransferase 5 in the colon
- PMID: 14534330
- PMCID: PMC240706
- DOI: 10.1073/pnas.2134464100
An endogenous retroviral long terminal repeat is the dominant promoter for human beta1,3-galactosyltransferase 5 in the colon
Abstract
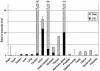
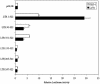
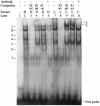
LTRs of endogenous retroviruses are known to affect expression of several human genes, typically as a relatively minor alternative promoter. Here, we report that an endogenous retrovirus LTR acts as one of at least two alternative promoters for the human beta1,3-galactosyltransferase 5 gene, involved in type 1 Lewis antigen synthesis, and show that the LTR promoter is most active in the gastrointestinal tract and mammary gland. Indeed, the LTR is the dominant promoter in the colon, indicating that this ancient retroviral element has a major impact on gene expression. Using colorectal cancer cell lines and electrophoretic mobility-shift assays, we found that hepatocyte nuclear factor 1 (HNF-1) binds a site within the retroviral promoter and that expression of HNF-1 and interaction with its binding site correlated with promoter activation. We conclude that HNF-1 is at least partially responsible for the tissue-specific activation of the LTR promoter of human beta 1,3-galactosyltransferase 5. We demonstrate that this tissue-specific transcription factor is implicated in the activation of an LTR gene promoter.
Figures





Similar articles
-
Endogenous retrovirus long terminal repeats as ready-to-use mobile promoters: the case of primate beta3GAL-T5.Gene. 2005 Dec 30;364:2-12. doi: 10.1016/j.gene.2005.05.045. Epub 2005 Aug 22. Gene. 2005. PMID: 16112824
-
Transcriptional control of the HERV-H LTR element of the GSDML gene in human tissues and cancer cells.Arch Virol. 2006 Oct;151(10):1985-94. doi: 10.1007/s00705-006-0764-5. Epub 2006 Apr 20. Arch Virol. 2006. PMID: 16625320
-
Cell type-specific expression and promoter activity of human endogenous retroviral long terminal repeats.Virology. 2001 Jan 5;279(1):280-91. doi: 10.1006/viro.2000.0712. Virology. 2001. PMID: 11145909
-
Endogenous retroviral LTRs as promoters for human genes: a critical assessment.Gene. 2009 Dec 15;448(2):105-14. doi: 10.1016/j.gene.2009.06.020. Epub 2009 Jul 3. Gene. 2009. PMID: 19577618 Review.
-
The role of human endogenous retroviral long terminal repeat sequences in human cancer (Review).Int J Mol Med. 2013 Oct;32(4):755-62. doi: 10.3892/ijmm.2013.1460. Epub 2013 Jul 30. Int J Mol Med. 2013. PMID: 23900638 Review.
Cited by
-
Microarray-based sketches of the HERV transcriptome landscape.PLoS One. 2012;7(6):e40194. doi: 10.1371/journal.pone.0040194. Epub 2012 Jun 28. PLoS One. 2012. PMID: 22761958 Free PMC article.
-
Prolactin in man: a tale of two promoters.Bioessays. 2006 Oct;28(10):1051-5. doi: 10.1002/bies.20468. Bioessays. 2006. PMID: 16998840 Free PMC article. Review.
-
Cis-regulatory elements in the Accord retrotransposon result in tissue-specific expression of the Drosophila melanogaster insecticide resistance gene Cyp6g1.Genetics. 2007 Mar;175(3):1071-7. doi: 10.1534/genetics.106.066597. Epub 2006 Dec 18. Genetics. 2007. PMID: 17179088 Free PMC article.
-
SSEA3 and Sialyl Lewis a Glycan Expression Is Controlled by B3GALT5 LTR through Lamin A-NFYA and SIRT1-STAT3 Signaling in Human ES Cells.Cells. 2020 Jan 10;9(1):177. doi: 10.3390/cells9010177. Cells. 2020. PMID: 31936807 Free PMC article.
-
Transposable elements in cancer as a by-product of stress-induced evolvability.Front Genet. 2014 May 30;5:156. doi: 10.3389/fgene.2014.00156. eCollection 2014. Front Genet. 2014. PMID: 24910642 Free PMC article.
References
-
- International Human Genome Sequencing Consortium (2001) Nature 409 860–921. - PubMed
-
- Bock, M. & Stoye, J. P. (2000) Curr. Opin. Genet. Dev. 10 651–655. - PubMed
-
- Stoye, J. P. (2001) Curr. Biol. 11 R914–R916. - PubMed
-
- Landry, J. R. & Mager, D. L. (2002) Genomics 80 499–508. - PubMed
-
- Medstrand, P., Landry, J. R. & Mager, D. L. (2001) J. Biol. Chem. 276 1896–1903. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

