HCF-1 functions as a coactivator for the zinc finger protein Krox20
- PMID: 14532282
- PMCID: PMC4291123
- DOI: 10.1074/jbc.M303470200
HCF-1 functions as a coactivator for the zinc finger protein Krox20
Abstract
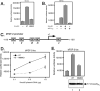
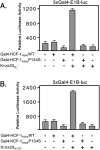
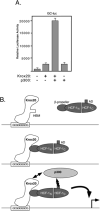
HCF-1 is a transcriptional cofactor required for activation of herpes simplex virus immediate-early genes by VP16 as well as less clearly defined roles in cell proliferation, cytokinesis, and spliceosome formation. It is expressed as a large precursor that undergoes proteolysis to yield two subunits that remain stably associated. VP16 uses a degenerate 4-amino acid sequence, known as the HCF-binding motif, to bind to a six-bladed beta-propeller domain at the N terminus of HCF-1. Functional HCF-binding motifs are also found in LZIP and Zhangfei, two cellular bZIP transcription factors of unknown function. Here we show that the HCF-binding motif occurs in a wide spectrum of DNA-binding proteins and transcriptional cofactors. Three well characterized examples were further analyzed for their ability to use HCF-1 as a coactivator. Krox20, a zinc finger transcription factor required for Schwann cell differentiation, and E2F4, a cell cycle regulator, showed a strong requirement for functional HCF-1 to activate transcription. In contrast, activation by estrogen receptor-alpha did not display HCF dependence. In Krox20, the HCF-binding motif lies within the N-terminal activation domain and mutation of this sequence diminishes both transactivation and association with the HCF-1 beta-propeller. The activation domain in the C-terminal subunit of HCF-1 contributes to activation by Krox20, possibly through recruitment of p300. These results suggest that HCF-1 is recruited by many different classes of cellular transcription factors and is therefore likely to be required for a variety of cellular processes including cell cycle progression and development.
Figures



Similar articles
-
N-terminal transcriptional activation domain of LZIP comprises two LxxLL motifs and the host cell factor-1 binding motif.Proc Natl Acad Sci U S A. 2000 Sep 26;97(20):10757-62. doi: 10.1073/pnas.190062797. Proc Natl Acad Sci U S A. 2000. PMID: 10984507 Free PMC article.
-
Mutations in host cell factor 1 separate its role in cell proliferation from recruitment of VP16 and LZIP.Mol Cell Biol. 2000 Feb;20(3):919-28. doi: 10.1128/MCB.20.3.919-928.2000. Mol Cell Biol. 2000. PMID: 10629049 Free PMC article.
-
Zhangfei is a potent and specific inhibitor of the host cell factor-binding transcription factor Luman.J Biol Chem. 2005 Apr 15;280(15):15257-66. doi: 10.1074/jbc.M500728200. Epub 2005 Feb 9. J Biol Chem. 2005. PMID: 15705566
-
Role of host cell factor-1 in cell cycle regulation.Transcription. 2012 Jul-Aug;3(4):187-92. doi: 10.4161/trns.20711. Epub 2012 Jul 1. Transcription. 2012. PMID: 22771988 Free PMC article. Review.
-
The herpes simplex virus VP16-induced complex: the makings of a regulatory switch.Trends Biochem Sci. 2003 Jun;28(6):294-304. doi: 10.1016/S0968-0004(03)00088-4. Trends Biochem Sci. 2003. PMID: 12826401 Review.
Cited by
-
The molecular machinery of myelin gene transcription in Schwann cells.Glia. 2008 Nov 1;56(14):1541-1551. doi: 10.1002/glia.20767. Glia. 2008. PMID: 18803322 Free PMC article. Review.
-
The dynamics of HCF-1 modulation of herpes simplex virus chromatin during initiation of infection.Viruses. 2013 May 22;5(5):1272-91. doi: 10.3390/v5051272. Viruses. 2013. PMID: 23698399 Free PMC article. Review.
-
The deubiquitinating enzyme BAP1 regulates cell growth via interaction with HCF-1.J Biol Chem. 2009 Dec 4;284(49):34179-88. doi: 10.1074/jbc.M109.046755. Epub 2009 Oct 8. J Biol Chem. 2009. PMID: 19815555 Free PMC article.
-
The THAP-zinc finger protein THAP1 associates with coactivator HCF-1 and O-GlcNAc transferase: a link between DYT6 and DYT3 dystonias.J Biol Chem. 2010 Apr 30;285(18):13364-71. doi: 10.1074/jbc.M109.072579. Epub 2010 Mar 3. J Biol Chem. 2010. PMID: 20200153 Free PMC article.
-
Genome-wide association identifies genomic regions influencing fillet color in Northwest Atlantic salmon (Salmo salar Linnaeus 1758).Front Genet. 2024 Jul 26;15:1402927. doi: 10.3389/fgene.2024.1402927. eCollection 2024. Front Genet. 2024. PMID: 39130751 Free PMC article.
References
-
- Kristie TM, Pomerantz JL, Twomey TC, Parent SA, Sharp PA. J Biol Chem. 1995;270:4387–4394. - PubMed
-
- Wilson AC, LaMarco K, Peterson MG, Herr W. Cell. 1993;74:115–125. - PubMed
-
- Wilson AC, Peterson MG, Herr W. Genes Dev. 1995;9:2445–2458. - PubMed
-
- Frattini A, Faranda S, Redolfi E, Zucchi I, Villa A, Patrosso MC, Strina D, Susani L, Vezzoni P. Genomics. 1994;23:30–35. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

