Single nucleotide variation analysis in 65 candidate genes for CNS disorders in a representative sample of the European population
- PMID: 14525928
- PMCID: PMC403700
- DOI: 10.1101/gr.1299703
Single nucleotide variation analysis in 65 candidate genes for CNS disorders in a representative sample of the European population
Abstract
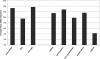
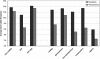
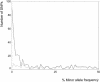
The detailed investigation of variation in functionally important regions of the human genome is expected to promote understanding of genetically complex diseases. We resequenced 65 candidate genes for CNS disorders in an average of 85 European individuals. The minor allele frequency (MAF), an indicator of weak purifying selection, was lowest in radical amino acid alterations, whereas similar MAF was observed for synonymous variants and conservative amino acid alterations. In noncoding sequences, variants located in CpG islands tended to have a lower MAF than those outside CpG islands. The transition/transversion ratio was increased among both synonymous and conservative variants compared with noncoding variants. Conversely, the transition/transversion ratio was lowest among radical amino acid alterations. Furthermore, among nonsynonymous variants, transversions displayed lower MAF than did transitions. This suggests that transversions are associated with functionally important amino acid alterations. By comparing our data with public SNP databases, we found that variants with lower allele frequency are underrepresented in these databases. Therefore, radical variants obtain distinctively lower database coverage. However, those variants appear to be under weak purifying selection and thus could play a role in the etiology of genetically complex diseases.
Figures
Similar articles
-
Systematic investigation of genetic variability in 111 human genes-implications for studying variable drug response.Pharmacogenomics J. 2005;5(3):183-92. doi: 10.1038/sj.tpj.6500306. Pharmacogenomics J. 2005. PMID: 15809674
-
Survey of polymorphic sequence variation in the immediate 5' region of human DNA repair genes.Mutat Res. 2007 Mar 1;616(1-2):221-6. doi: 10.1016/j.mrfmmm.2006.11.008. Epub 2006 Dec 11. Mutat Res. 2007. PMID: 17161851
-
Towards realistic codon models: among site variability and dependency of synonymous and non-synonymous rates.Bioinformatics. 2007 Jul 1;23(13):i319-27. doi: 10.1093/bioinformatics/btm176. Bioinformatics. 2007. PMID: 17646313
-
SNPs: impact on gene function and phenotype.Methods Mol Biol. 2009;578:3-22. doi: 10.1007/978-1-60327-411-1_1. Methods Mol Biol. 2009. PMID: 19768584 Review.
-
Gene variants in noncoding regions and their possible consequences.Pharmacogenomics. 2006 Mar;7(2):203-9. doi: 10.2217/14622416.7.2.203. Pharmacogenomics. 2006. PMID: 16515399 Review.
Cited by
-
Large-scale characterization of public database SNPs causing non-synonymous changes in three ethnic groups.Hum Genet. 2006 Mar;119(1-2):75-83. doi: 10.1007/s00439-005-0105-x. Epub 2005 Dec 14. Hum Genet. 2006. PMID: 16391945
-
Allele balance bias identifies systematic genotyping errors and false disease associations.Hum Mutat. 2019 Jan;40(1):115-126. doi: 10.1002/humu.23674. Epub 2018 Nov 23. Hum Mutat. 2019. PMID: 30353964 Free PMC article.
-
Establishment of a pipeline to analyse non-synonymous SNPs in Bos taurus.BMC Genomics. 2006 Nov 26;7:298. doi: 10.1186/1471-2164-7-298. BMC Genomics. 2006. PMID: 17125523 Free PMC article.
-
Analyses of porcine public SNPs in coding-gene regions by re-sequencing and phenotypic association studies.Mol Biol Rep. 2011 Aug;38(6):3805-20. doi: 10.1007/s11033-010-0496-1. Epub 2010 Nov 24. Mol Biol Rep. 2011. PMID: 21107721
-
Targeted sequencing of both DNA strands barcoded and captured individually by RNA probes to identify genome-wide ultra-rare mutations.Sci Rep. 2017 Jun 13;7(1):3356. doi: 10.1038/s41598-017-03448-8. Sci Rep. 2017. PMID: 28611392 Free PMC article.
References
-
- Botstein, D. and Risch, N. 2003. Discovering genotypes underlying human phenotypes: Past successes for mendelian disease, future approaches for complex disease. Nat. Genet. 33: 228-237. - PubMed
-
- Cargill, M., Altshuler, D., Ireland, J., Sklar, P., Ardlie, K., Patil, N., Shaw, N., Lane, C.R., Lim, E.P., Kalyanaraman, N., et al. 1999. Characterization of single-nucleotide polymorphisms in coding regions of human genes. Nat. Genet. 22: 231-238. - PubMed
-
- Carlson, C.S., Eberle, M.L., Rieder, M.J., Smith, J.D., Kruglyak, L., and Nickerson, D.A. 2003. Additional SNPs and linkage-disequilibrium analyses are necessary for whole-genome association studies in humans. Nat. Genet. 33: 518-521. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
