In vitro proteolytic processing of the MD145 norovirus ORF1 nonstructural polyprotein yields stable precursors and products similar to those detected in calicivirus-infected cells
- PMID: 14512545
- PMCID: PMC224964
- DOI: 10.1128/jvi.77.20.10957-10974.2003
In vitro proteolytic processing of the MD145 norovirus ORF1 nonstructural polyprotein yields stable precursors and products similar to those detected in calicivirus-infected cells
Abstract
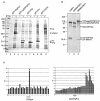
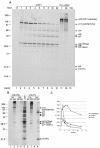
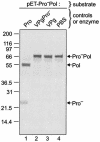
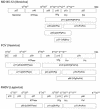
The MD145-12 strain (GII/4) is a member of the genus Norovirus in the Caliciviridae and was detected in a patient with acute gastroenteritis in a Maryland nursing home. The open reading frame 1 (ORF1) (encoding the nonstructural polyprotein) was cloned as a consensus sequence into various expression vectors, and a proteolytic cleavage map was determined. The virus-encoded cysteine proteinase mediated at least five cleavages (Q(330)/G(331), Q(696)/G(697), E(875)/G(876), E(1008)/A(1009), and E(1189)/G(1190)) in the ORF1 polyprotein in the following order: N-terminal protein; nucleoside triphosphatase; 20-kDa protein (p20); virus protein, genome linked (VPg); proteinase (Pro); polymerase (Pol). A time course analysis of proteolytic processing of the MD145-12 ORF1 polyprotein in an in vitro coupled transcription and translation assay allowed the identification of stable precursors and final mapped cleavage products. Stable precursors included p20VPg (analogous to the 3AB of the picornaviruses) and ProPol (analogous to the 3CD of the picornaviruses). Less stable processing intermediates were identified as p20VPgProPol, p20VPgPro, and VPgPro. The MD145-12 Pro and ProPol proteins were expressed in bacteria as active forms of the proteinase and used to further characterize their substrate specificities in trans cleavage assays. The MD145-12 Pro was able to cleave its five mapped cleavage sites in trans and, in addition, could mediate trans cleavage of the Norwalk virus (GI/I) ORF1 polyprotein into a similar proteolytic processing profile. Taken together, our data establish a model for proteolytic processing in the noroviruses that is consistent with nonstructural precursors and products identified in studies of caliciviruses that replicate in cell culture systems.
Figures





Similar articles
-
Processing map and essential cleavage sites of the nonstructural polyprotein encoded by ORF1 of the feline calicivirus genome.J Virol. 2002 Jul;76(14):7060-72. doi: 10.1128/jvi.76.14.7060-7072.2002. J Virol. 2002. PMID: 12072506 Free PMC article.
-
Proteolytic processing of sapovirus ORF1 polyprotein.J Virol. 2005 Jun;79(12):7283-90. doi: 10.1128/JVI.79.12.7283-7290.2005. J Virol. 2005. PMID: 15919882 Free PMC article.
-
The p4-p2' amino acids surrounding human norovirus polyprotein cleavage sites define the core sequence regulating self-processing order.J Virol. 2014 Sep;88(18):10738-47. doi: 10.1128/JVI.01357-14. Epub 2014 Jul 2. J Virol. 2014. PMID: 24991013 Free PMC article.
-
[Genomic organization of Norwalk-like viruses and functions of viral gene products].Nihon Rinsho. 2002 Jun;60(6):1155-64. Nihon Rinsho. 2002. PMID: 12078089 Review. Japanese.
-
Prokaryotic polyprotein precursors.FEBS Lett. 1992 Jul 27;307(1):62-5. doi: 10.1016/0014-5793(92)80902-s. FEBS Lett. 1992. PMID: 1639196 Review.
Cited by
-
Structure determination of Murine Norovirus NS6 proteases with C-terminal extensions designed to probe protease-substrate interactions.PeerJ. 2015 Feb 26;3:e798. doi: 10.7717/peerj.798. eCollection 2015. PeerJ. 2015. PMID: 25755927 Free PMC article.
-
Structure of a murine norovirus NS6 protease-product complex revealed by adventitious crystallisation.PLoS One. 2012;7(6):e38723. doi: 10.1371/journal.pone.0038723. Epub 2012 Jun 7. PLoS One. 2012. PMID: 22685603 Free PMC article.
-
Comparison of the replication properties of murine and human calicivirus RNA-dependent RNA polymerases.Virus Genes. 2011 Feb;42(1):16-27. doi: 10.1007/s11262-010-0535-y. Epub 2010 Oct 20. Virus Genes. 2011. PMID: 20960046
-
Model systems for the study of human norovirus Biology.Future Virol. 2009 Jul;4(4):353-367. doi: 10.2217/fvl.09.18. Future Virol. 2009. PMID: 21516251 Free PMC article.
-
Contribution of intra- and interhost dynamics to norovirus evolution.J Virol. 2012 Mar;86(6):3219-29. doi: 10.1128/JVI.06712-11. Epub 2011 Dec 28. J Virol. 2012. PMID: 22205753 Free PMC article.
References
-
- Ando, T., J. S. Noel, and R. L. Fankhauser. 2000. Genetic classification of “Norwalk-like viruses.” J. Infect. Dis. 181(Suppl. 2):S336-S348. - PubMed
-
- Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

