Ectopic expression of atRSZ33 reveals its function in splicing and causes pleiotropic changes in development
- PMID: 12972547
- PMCID: PMC196550
- DOI: 10.1091/mbc.e03-02-0109
Ectopic expression of atRSZ33 reveals its function in splicing and causes pleiotropic changes in development
Abstract
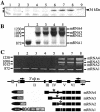
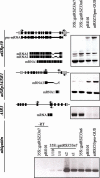
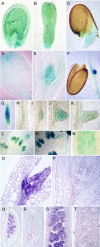
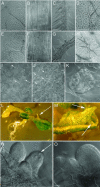
Splicing provides an additional level in the regulation of gene expression and contributes to proteome diversity. Herein, we report the functional characterization of a recently described plant-specific protein, atRSZ33, which has characteristic features of a serine/arginine-rich protein and the ability to interact with other splicing factors, implying that this protein might be involved in constitutive and/or alternative splicing. Overexpression of atRSZ33 leads to alteration of splicing patterns of atSRp30 and atSRp34/SR1, indicating that atRSZ33 is indeed a splicing factor. Moreover, atRSZ33 is a regulator of its own expression, as splicing of its pre-mRNA is changed in transgenic plants. Investigations by promoter-beta-glucuronidase (GUS) fusion and in situ hybridization revealed that atRSZ33 is expressed during embryogenesis and early stages of seedling formation, as well as in flower and root development. Ectopic expression of atRSZ33 caused pleiotropic changes in plant development resulting in increased cell expansion and changed polarization of cell elongation and division. In addition, changes in activity of an auxin-responsive promoter suggest that auxin signaling is disturbed in these transgenic plants.
Figures

Similar articles
-
atSRp30, one of two SF2/ASF-like proteins from Arabidopsis thaliana, regulates splicing of specific plant genes.Genes Dev. 1999 Apr 15;13(8):987-1001. doi: 10.1101/gad.13.8.987. Genes Dev. 1999. PMID: 10215626 Free PMC article.
-
Network of interactions of a novel plant-specific Arg/Ser-rich protein, atRSZ33, with atSC35-like splicing factors.J Biol Chem. 2002 Oct 18;277(42):39989-98. doi: 10.1074/jbc.M206455200. Epub 2002 Aug 9. J Biol Chem. 2002. PMID: 12176998
-
The auxin-regulated AP2/EREBP gene PUCHI is required for morphogenesis in the early lateral root primordium of Arabidopsis.Plant Cell. 2007 Jul;19(7):2156-68. doi: 10.1105/tpc.107.050674. Epub 2007 Jul 13. Plant Cell. 2007. PMID: 17630277 Free PMC article.
-
[Mechanisms of alternative splicing in regulating plant flowering: a review].Sheng Wu Gong Cheng Xue Bao. 2021 Sep 25;37(9):2991-3004. doi: 10.13345/j.cjb.200628. Sheng Wu Gong Cheng Xue Bao. 2021. PMID: 34622612 Review. Chinese.
-
Arabidopsis embryogenesis. Radicle development(s).Curr Biol. 1995 Feb 1;5(2):110-2. doi: 10.1016/s0960-9822(95)00027-3. Curr Biol. 1995. PMID: 7743170 Review.
Cited by
-
Regulatory Network of Serine/Arginine-Rich (SR) Proteins: The Molecular Mechanism and Physiological Function in Plants.Int J Mol Sci. 2022 Sep 5;23(17):10147. doi: 10.3390/ijms231710147. Int J Mol Sci. 2022. PMID: 36077545 Free PMC article. Review.
-
Abscisic acid (ABA) regulation of Arabidopsis SR protein gene expression.Int J Mol Sci. 2014 Sep 29;15(10):17541-64. doi: 10.3390/ijms151017541. Int J Mol Sci. 2014. PMID: 25268622 Free PMC article.
-
The Arabidopsis SR45 Splicing Factor, a Negative Regulator of Sugar Signaling, Modulates SNF1-Related Protein Kinase 1 Stability.Plant Cell. 2016 Aug;28(8):1910-25. doi: 10.1105/tpc.16.00301. Epub 2016 Jul 19. Plant Cell. 2016. PMID: 27436712 Free PMC article.
-
Maize rough endosperm3 encodes an RNA splicing factor required for endosperm cell differentiation and has a nonautonomous effect on embryo development.Plant Cell. 2011 Dec;23(12):4280-97. doi: 10.1105/tpc.111.092163. Epub 2011 Dec 2. Plant Cell. 2011. PMID: 22138152 Free PMC article.
-
Imaging of endogenous messenger RNA splice variants in living cells reveals nuclear retention of transcripts inaccessible to nonsense-mediated decay in Arabidopsis.Plant Cell. 2014 Feb;26(2):754-64. doi: 10.1105/tpc.113.118075. Epub 2014 Feb 14. Plant Cell. 2014. PMID: 24532591 Free PMC article.
References
-
- Barta, A., Sommergruber, K., Thompson, D., Hartmuth, K., Matzke, M.A., and Matzke, A.J.M. (1986). The expression of a nopaline synthase-human growth hormone chimaeric gene in transformed tobacco and sunflower callus tissue. Plant Mol. Biol. 6, 347–357. - PubMed
-
- Baskin, T.I., Busby, C.H., Fowke, L.C., Sammut, M., and Gubler, E. (1992). Improvements in immunostaining samples embedded in methacrylate: localization of microtubules and other antigens throughout developing organs in plants of diverse taxa. Planta 187, 405–413. - PubMed
-
- Bennett, M.J., Marchant, A., Green, H.G., May, S.T., Ward, S.P., Millner, P.A., Walker, A.R., Schulz, B., and Feldmann, K.A. (1996). Arabidopsis AUX1 gene: a permease-like regulator of root gravitropism. Science 273, 948–950. - PubMed
-
- Caceres, J.F., and Krainer, A.R. (1997). Mammalian pre-mRNA splicing factors. In: Eukaryotic mRNA Processing: Frontiers in Molecular Biology, ed. A.R. Krainer, Oxford: IRL Press, 174–212.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

