BRITTLE CULM1, which encodes a COBRA-like protein, affects the mechanical properties of rice plants
- PMID: 12953108
- PMCID: PMC181328
- DOI: 10.1105/tpc.011775
BRITTLE CULM1, which encodes a COBRA-like protein, affects the mechanical properties of rice plants
Abstract
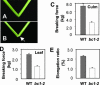
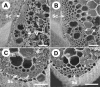
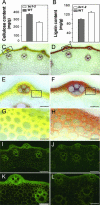
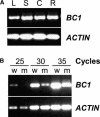
Plant mechanical strength is an important agronomic trait. To understand the molecular mechanism that controls the plant mechanical strength of crops, we characterized the classic rice mutant brittle culm1 (bc1) and isolated BC1 using a map-based cloning approach. BC1, which encodes a COBRA-like protein, is expressed mainly in developing sclerenchyma cells and in vascular bundles of rice. In these types of cells, mutations in BC1 cause not only a reduction in cell wall thickness and cellulose content but also an increase in lignin level, suggesting that BC1, a gene that controls the mechanical strength of monocots, plays an important role in the biosynthesis of the cell walls of mechanical tissues.
Figures



Similar articles
-
Mutation of rice bc1 gene affects internode elongation and induces delayed cell wall deposition in developing internodes.Plant Signal Behav. 2020 May 3;15(5):1749786. doi: 10.1080/15592324.2020.1749786. Epub 2020 Apr 16. Plant Signal Behav. 2020. PMID: 32299283 Free PMC article.
-
Maize Brittle stalk2 encodes a COBRA-like protein expressed in early organ development but required for tissue flexibility at maturity.Plant Physiol. 2007 Dec;145(4):1444-59. doi: 10.1104/pp.107.102582. Epub 2007 Oct 11. Plant Physiol. 2007. PMID: 17932309 Free PMC article.
-
Rice BRITTLE CULM 5 (BRITTLE NODE) is involved in secondary cell wall formation in the sclerenchyma tissue of nodes.Plant Cell Physiol. 2009 Nov;50(11):1886-97. doi: 10.1093/pcp/pcp133. Epub 2009 Oct 6. Plant Cell Physiol. 2009. PMID: 19812064 Free PMC article.
-
Brittle Culm 1 Encodes a COBRA-Like Protein Involved in Secondary Cell Wall Cellulose Biosynthesis in Sorghum.Plant Cell Physiol. 2019 Apr 1;60(4):788-801. doi: 10.1093/pcp/pcy246. Plant Cell Physiol. 2019. PMID: 30590744
-
A missense mutation in the transmembrane domain of CESA9 affects cell wall biosynthesis and plant growth in rice.Plant Sci. 2012 Nov;196:117-24. doi: 10.1016/j.plantsci.2012.08.002. Epub 2012 Aug 9. Plant Sci. 2012. PMID: 23017906
Cited by
-
Genome-Wide Association Study Identified Novel Candidate Loci/Genes Affecting Lodging Resistance in Rice.Genes (Basel). 2021 May 11;12(5):718. doi: 10.3390/genes12050718. Genes (Basel). 2021. PMID: 34064770 Free PMC article.
-
CEF3 is involved in membrane trafficking and essential for secondary cell wall biosynthesis and its mutation enhanced biomass enzymatic saccharification in rice.Biotechnol Biofuels Bioprod. 2022 Oct 14;15(1):111. doi: 10.1186/s13068-022-02205-y. Biotechnol Biofuels Bioprod. 2022. PMID: 36242043 Free PMC article.
-
Predicting the size of the progeny mapping population required to positionally clone a gene.Genetics. 2007 Aug;176(4):2035-54. doi: 10.1534/genetics.107.074377. Epub 2007 Jun 11. Genetics. 2007. PMID: 17565938 Free PMC article.
-
Combining expression and comparative evolutionary analysis. The COBRA gene family.Plant Physiol. 2007 Jan;143(1):172-87. doi: 10.1104/pp.106.087262. Epub 2006 Nov 10. Plant Physiol. 2007. PMID: 17098858 Free PMC article.
-
Phenotypic plasticity in cell walls of maize brown midrib mutants is limited by lignin composition.J Exp Bot. 2010 May;61(9):2479-90. doi: 10.1093/jxb/erq093. Epub 2010 Apr 21. J Exp Bot. 2010. PMID: 20410320 Free PMC article.
References
-
- Arioli, T., et al. (1998). Molecular analysis of cellulose biosynthesis in Arabidopsis. Science 279, 717–720. - PubMed
-
- Carpita, N., and McCann, M. (2000). The cell wall. In Biochemistry and Molecular Biology of Plants, B.B. Buchanan, W. Gruissem, and R.L. Jones, eds (Rockville, MD: American Society of Plant Physiologists), pp. 52–108.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

