The human I-mfa domain-containing protein, HIC, interacts with cyclin T1 and modulates P-TEFb-dependent transcription
- PMID: 12944466
- PMCID: PMC193714
- DOI: 10.1128/MCB.23.18.6373-6384.2003
The human I-mfa domain-containing protein, HIC, interacts with cyclin T1 and modulates P-TEFb-dependent transcription
Abstract
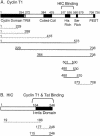
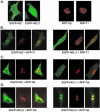
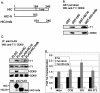
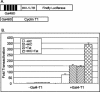
Positive transcription elongation factor b (P-TEFb) hyperphosphorylates the carboxy-terminal domain of RNA polymerase II, permitting productive transcriptional elongation. The cyclin T1 subunit of P-TEFb engages cellular transcription factors as well as the human immunodeficiency virus type 1 (HIV-1) transactivator Tat. To identify potential P-TEFb regulators, we conducted a yeast two-hybrid screen with cyclin T1 as bait. Among the proteins isolated was the human I-mfa domain-containing protein (HIC). HIC has been reported to modulate expression from both cellular and viral promoters via its C-terminal cysteine-rich domain, which is similar to the inhibitor of MyoD family a (I-mfa) protein. We show that HIC binds cyclin T1 in yeast and mammalian cells and that it interacts with intact P-TEFb in mammalian cell extracts. The interaction involves the I-mfa domain of HIC and the regulatory histidine-rich region of cyclin T1. HIC also binds Tat via its I-mfa domain, although the sequence requirements are different. HIC colocalizes with cyclin T1 in nuclear speckle regions and with Tat in the nucleolus. Expression of the HIC cDNA modulates Tat transactivation of the HIV-1 long terminal repeat (LTR) in a cell type-specific fashion. It is mildly inhibitory in CEM cells but stimulates gene expression in HeLa, COS, and NIH 3T3 cells. The isolated I-mfa domain acts as a dominant negative inhibitor. Activation of the HIV-1 LTR by HIC in NIH 3T3 cells occurs at the RNA level and is mediated by direct interactions with P-TEFb.
Figures



Similar articles
-
Developmental regulators containing the I-mfa domain interact with T cyclins and Tat and modulate transcription.J Mol Biol. 2007 Mar 30;367(3):630-46. doi: 10.1016/j.jmb.2007.01.020. Epub 2007 Jan 12. J Mol Biol. 2007. PMID: 17289077 Free PMC article.
-
Recruitment of cyclin T1/P-TEFb to an HIV type 1 long terminal repeat promoter proximal RNA target is both necessary and sufficient for full activation of transcription.Proc Natl Acad Sci U S A. 1999 Jul 6;96(14):7791-6. doi: 10.1073/pnas.96.14.7791. Proc Natl Acad Sci U S A. 1999. PMID: 10393900 Free PMC article.
-
The growth factor granulin interacts with cyclin T1 and modulates P-TEFb-dependent transcription.Mol Cell Biol. 2003 Mar;23(5):1688-702. doi: 10.1128/MCB.23.5.1688-1702.2003. Mol Cell Biol. 2003. PMID: 12588988 Free PMC article.
-
Regulatory functions of Cdk9 and of cyclin T1 in HIV tat transactivation pathway gene expression.J Cell Biochem. 1999 Dec 1;75(3):357-68. J Cell Biochem. 1999. PMID: 10536359 Review.
-
How the sequestration of a protein interferes with its mechanism of action: example of a new family of proteins characterized by a particular cysteine-rich carboxy-terminal domain involved in gene expression regulation.Curr Protein Pept Sci. 2001 Jun;2(2):155-67. doi: 10.2174/1389203013381143. Curr Protein Pept Sci. 2001. PMID: 12370022 Review.
Cited by
-
Dominant negative mutant cyclin T1 proteins inhibit HIV transcription by specifically degrading Tat.Retrovirology. 2008 Jul 11;5:63. doi: 10.1186/1742-4690-5-63. Retrovirology. 2008. PMID: 18620576 Free PMC article.
-
Differential host cell gene expression and regulation of cell cycle progression by nonstructural protein 11 of porcine reproductive and respiratory syndrome virus.Biomed Res Int. 2014;2014:430508. doi: 10.1155/2014/430508. Epub 2014 Feb 26. Biomed Res Int. 2014. PMID: 24719865 Free PMC article.
-
The role of Rhox homeobox factors in tumorigenesis.Front Biosci (Landmark Ed). 2013 Jan 1;18(2):474-92. doi: 10.2741/4115. Front Biosci (Landmark Ed). 2013. PMID: 23276937 Free PMC article. Review.
-
The complex regulation of HIC (Human I-mfa domain containing protein) expression.PLoS One. 2009 Jul 7;4(7):e6152. doi: 10.1371/journal.pone.0006152. PLoS One. 2009. PMID: 19582149 Free PMC article.
-
The Yin and Yang of P-TEFb regulation: implications for human immunodeficiency virus gene expression and global control of cell growth and differentiation.Microbiol Mol Biol Rev. 2006 Sep;70(3):646-59. doi: 10.1128/MMBR.00011-06. Microbiol Mol Biol Rev. 2006. PMID: 16959964 Free PMC article. Review.
References
-
- Barboric, M., R. M. Nissen, S. Kanazawa, N. Jabrane-Ferrat, and B. M. Peterlin. 2001. NF-kappaB binds P-TEFb to stimulate transcriptional elongation by RNA polymerase II. Mol. Cell 8:327-337. - PubMed
-
- Chen, C. M., N. Kraut, M. Groudine, and H. Weintraub. 1996. I-mf, a novel myogenic repressor, interacts with members of the MyoD family. Cell 86:731-741. - PubMed
-
- Darbinian, N., B. E. Sawaya, K. Khalili, N. Jaffe, B. Wortman, A. Giordano, and S. Amini. 2001. Functional interaction between cyclin T1/cdk9 and Puralpha determines the level of TNFalpha promoter activation by Tat in glial cells. J. Neuroimmunol. 121:3-11. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
