Gangliosides are receptors for murine polyoma virus and SV40
- PMID: 12941687
- PMCID: PMC202381
- DOI: 10.1093/emboj/cdg439
Gangliosides are receptors for murine polyoma virus and SV40
Abstract
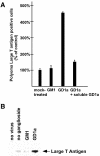
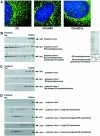
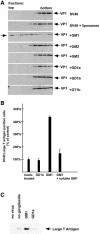
Polyoma virus (Py) and simian virus 40 (SV40) travel from the plasma membrane to the endoplasmic reticulum (ER) from where they enter the cytosol and then the nucleus to initiate infection. Here we demonstrate that specific gangliosides can serve as plasma membrane receptors for these viruses, GD1a and GT1b for Py and GM1 for SV40. Binding and flotation assays were used to show that addition of these gangliosides to phospholipid vesicles allowed specific binding of the respective viruses. The crystal structure of polyoma VP1 with a sialic acid-containing oligosaccharide was used to derive a model of how the two terminal sugars (sialic acid-alpha2,3-galactose) in one branch of GD1a and GT1b are recognized by the virus. A rat cell line deficient in ganglioside synthesis is poorly infectible by polyoma and SV40, but addition of the appropriate gangliosides greatly facilitates virus uptake, transport to the ER and infection. Lipid binding sites for polyoma are shown to be present in rough ER membranes, suggesting that the virus travel with the ganglioside(s) from the plasma membranes to the ER.
Figures


Similar articles
-
Identification of gangliosides GD1b and GT1b as receptors for BK virus.J Virol. 2006 Feb;80(3):1361-6. doi: 10.1128/JVI.80.3.1361-1366.2006. J Virol. 2006. PMID: 16415013 Free PMC article.
-
N-glycolyl GM1 ganglioside as a receptor for simian virus 40.J Virol. 2007 Dec;81(23):12846-58. doi: 10.1128/JVI.01311-07. Epub 2007 Sep 12. J Virol. 2007. PMID: 17855525 Free PMC article.
-
Uptake pathway of polyomavirus via ganglioside GD1a.J Virol. 2004 Nov;78(22):12259-67. doi: 10.1128/JVI.78.22.12259-12267.2004. J Virol. 2004. PMID: 15507613 Free PMC article.
-
Cellular entry of polyomaviruses.Curr Top Microbiol Immunol. 2010;343:177-94. doi: 10.1007/82_2010_38. Curr Top Microbiol Immunol. 2010. PMID: 20373089 Review.
-
Retrograde transport pathways utilised by viruses and protein toxins.Virol J. 2006 Apr 7;3:26. doi: 10.1186/1743-422X-3-26. Virol J. 2006. PMID: 16603059 Free PMC article. Review.
Cited by
-
Gangliosides are essential for bovine adeno-associated virus entry.J Virol. 2006 Jun;80(11):5516-22. doi: 10.1128/JVI.02393-05. J Virol. 2006. PMID: 16699032 Free PMC article.
-
Super-Resolution Microscopy Reveals Local Accumulation of Plasma Membrane Gangliosides at Neisseria meningitidis Invasion Sites.Front Cell Dev Biol. 2019 Sep 13;7:194. doi: 10.3389/fcell.2019.00194. eCollection 2019. Front Cell Dev Biol. 2019. PMID: 31572726 Free PMC article.
-
Infectious Entry of Merkel Cell Polyomavirus.J Virol. 2019 Mar 5;93(6):e02004-18. doi: 10.1128/JVI.02004-18. Print 2019 Mar 15. J Virol. 2019. PMID: 30626687 Free PMC article.
-
How non-enveloped viruses hijack host machineries to cause infection.Adv Virus Res. 2019;104:97-122. doi: 10.1016/bs.aivir.2019.05.002. Epub 2019 Jul 2. Adv Virus Res. 2019. PMID: 31439154 Free PMC article.
-
JC Polyomavirus Uses Extracellular Vesicles To Infect Target Cells.mBio. 2019 Apr 9;10(2):e00379-19. doi: 10.1128/mBio.00379-19. mBio. 2019. PMID: 30967463 Free PMC article.
References
-
- Anderson H.A., Chen,Y. and Norkin,L.C. (1998) MHC class I molecules are enriched in caveolae but do not enter with simian virus 40. J. Gen. Virol., 79, 1469–1477. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

