Activation of heat shock genes is not necessary for protection by heat shock transcription factor 1 against cell death due to a single exposure to high temperatures
- PMID: 12897157
- PMCID: PMC166333
- DOI: 10.1128/MCB.23.16.5882-5895.2003
Activation of heat shock genes is not necessary for protection by heat shock transcription factor 1 against cell death due to a single exposure to high temperatures
Abstract
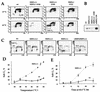
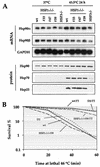
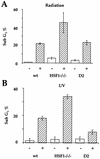
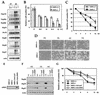
Heat shock response, which is characterized by the induction of a set of heat shock proteins, is essential for induced thermotolerance and is regulated by heat shock transcription factors (HSFs). Curiously, HSF1 is essential for heat shock response in mammals, whereas in avian HSF3, an avian-specific factor is required for the burst activation of heat shock genes. Amino acid sequences of chicken HSF1 are highly conserved with human HSF1, but those of HSF3 diverge significantly. Here, we demonstrated that chicken HSF1 lost the ability to activate heat shock genes through the amino-terminal domain containing an alanine-rich sequence and a DNA-binding domain. Surprisingly, chicken and human HSF1 but not HSF3 possess a novel function that protects against a single exposure to mild heat shock, which is not mediated through the activation of heat shock genes. Overexpression of HSF1 mutants that could not bind to DNA did not restore the susceptibility to cell death in HSF1-null cells, suggesting that the new protective role of HSF1 is mediated through regulation of unknown target genes other than heat shock genes. These results uncover a novel role of vertebrate HSF1, which has been masked under the roles of heat shock proteins.
Figures




Similar articles
-
The DNA-binding properties of two heat shock factors, HSF1 and HSF3, are induced in the avian erythroblast cell line HD6.Mol Cell Biol. 1995 Oct;15(10):5268-78. doi: 10.1128/MCB.15.10.5268. Mol Cell Biol. 1995. PMID: 7565675 Free PMC article.
-
Disruption of the HSF3 gene results in the severe reduction of heat shock gene expression and loss of thermotolerance.EMBO J. 1998 Mar 16;17(6):1750-8. doi: 10.1093/emboj/17.6.1750. EMBO J. 1998. PMID: 9501096 Free PMC article.
-
Reciprocal activation of HSF1 and HSF3 in brain and blood tissues: is redundancy developmentally related?Am J Physiol Regul Integr Comp Physiol. 2006 Sep;291(3):R566-72. doi: 10.1152/ajpregu.00685.2005. Am J Physiol Regul Integr Comp Physiol. 2006. PMID: 16497816
-
Roles of the heat shock transcription factors in regulation of the heat shock response and beyond.FASEB J. 2001 May;15(7):1118-31. doi: 10.1096/fj00-0294rev. FASEB J. 2001. PMID: 11344080 Review.
-
The heat shock factor family and adaptation to proteotoxic stress.FEBS J. 2010 Oct;277(20):4112-25. doi: 10.1111/j.1742-4658.2010.07827.x. FEBS J. 2010. PMID: 20945528 Review.
Cited by
-
Impaired hippocampal spinogenesis and neurogenesis and altered affective behavior in mice lacking heat shock factor 1.Proc Natl Acad Sci U S A. 2011 Jan 25;108(4):1681-6. doi: 10.1073/pnas.1016424108. Epub 2011 Jan 4. Proc Natl Acad Sci U S A. 2011. PMID: 21205885 Free PMC article.
-
Antioxidant Defence Systems and Oxidative Stress in Poultry Biology: An Update.Antioxidants (Basel). 2019 Jul 22;8(7):235. doi: 10.3390/antiox8070235. Antioxidants (Basel). 2019. PMID: 31336672 Free PMC article. Review.
-
HSF1 is required for cellular adaptation to daily temperature fluctuations.Sci Rep. 2024 Sep 12;14(1):21361. doi: 10.1038/s41598-024-72415-x. Sci Rep. 2024. PMID: 39266731 Free PMC article.
-
Subcellular distribution of human RDM1 protein isoforms and their nucleolar accumulation in response to heat shock and proteotoxic stress.Nucleic Acids Res. 2007;35(19):6571-87. doi: 10.1093/nar/gkm753. Epub 2007 Sep 28. Nucleic Acids Res. 2007. PMID: 17905820 Free PMC article.
-
Heat Shock Related Protein Expression in Abdominal Testes of Asian Elephant (Elephas maximus).Animals (Basel). 2024 Jul 30;14(15):2211. doi: 10.3390/ani14152211. Animals (Basel). 2024. PMID: 39123737 Free PMC article.
References
-
- C. N. Adra, P. H. Boer, and M. W. McBurney. 1987. Cloning and expression of the mouse pgk-1 gene and the nucleotide sequence of its promoter. Gene 60:65-74. - PubMed
-
- Aschoff, J., and U. von Saint Paul. 1973. Brain temperature as related to gross motor activity in the unanesthetized chicken. Physiol. Behav. 10:529-533. - PubMed
-
- Beere, H. M., B. B. Wolf, K. Cain, et al. 2000. Heat-shock protein 70 inhibits apoptosis by preventing recruitment of procaspase-9 to the Apaf-1 apoptosome. Nat. Cell Biol. 2:469-475. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
