The cytoplasmic tails of infectious bronchitis virus E and M proteins mediate their interaction
- PMID: 12890618
- PMCID: PMC7127533
- DOI: 10.1016/s0042-6822(03)00175-2
The cytoplasmic tails of infectious bronchitis virus E and M proteins mediate their interaction
Abstract
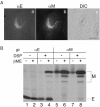
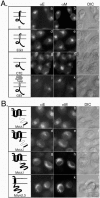
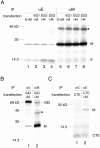
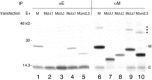
Virus-like particle (VLP) formation by the coronavirus E and M proteins suggests that interactions between these proteins play a critical role in coronavirus assembly. We studied interactions between the infectious bronchitis virus (IBV) E and M proteins using in vivo crosslinking and VLP assembly assays. We show that IBV E and M can be crosslinked to each other in IBV-infected and transfected cells, indicating that they interact. The cytoplasmic tails of both proteins are important for this interaction. We also examined the ability of the mutant and chimeric E and M proteins to form VLPs. IBV M proteins that are missing portions of their cytoplasmic tails or transmembrane regions were not able to support VLP formation, regardless of their ability to be crosslinked to IBV E. Interactions between the E and M proteins and the membrane bilayer are likely to play an important role in VLP formation and virus budding.
Figures



Similar articles
-
Infectious bronchitis virus E protein is targeted to the Golgi complex and directs release of virus-like particles.J Virol. 2000 May;74(9):4319-26. doi: 10.1128/jvi.74.9.4319-4326.2000. J Virol. 2000. PMID: 10756047 Free PMC article.
-
The Infectious Bronchitis Coronavirus Envelope Protein Alters Golgi pH To Protect the Spike Protein and Promote the Release of Infectious Virus.J Virol. 2019 May 15;93(11):e00015-19. doi: 10.1128/JVI.00015-19. Print 2019 Jun 1. J Virol. 2019. PMID: 30867314 Free PMC article.
-
A single polar residue and distinct membrane topologies impact the function of the infectious bronchitis coronavirus E protein.PLoS Pathog. 2012;8(5):e1002674. doi: 10.1371/journal.ppat.1002674. Epub 2012 May 3. PLoS Pathog. 2012. PMID: 22570613 Free PMC article.
-
Incorporation of spike and membrane glycoproteins into coronavirus virions.Viruses. 2015 Apr 3;7(4):1700-25. doi: 10.3390/v7041700. Viruses. 2015. PMID: 25855243 Free PMC article. Review.
-
The coronavirus E protein: assembly and beyond.Viruses. 2012 Mar;4(3):363-82. doi: 10.3390/v4030363. Epub 2012 Mar 8. Viruses. 2012. PMID: 22590676 Free PMC article. Review.
Cited by
-
Understanding COVID-19 via comparative analysis of dark proteomes of SARS-CoV-2, human SARS and bat SARS-like coronaviruses.Cell Mol Life Sci. 2021 Feb;78(4):1655-1688. doi: 10.1007/s00018-020-03603-x. Epub 2020 Jul 25. Cell Mol Life Sci. 2021. PMID: 32712910 Free PMC article.
-
Sequence and structural analysis of COVID-19 E and M proteins with MERS virus E and M proteins-A comparative study.Biochem Biophys Rep. 2021 Jul;26:101023. doi: 10.1016/j.bbrep.2021.101023. Epub 2021 May 14. Biochem Biophys Rep. 2021. PMID: 34013072 Free PMC article.
-
Biochemical evidence for the presence of mixed membrane topologies of the severe acute respiratory syndrome coronavirus envelope protein expressed in mammalian cells.FEBS Lett. 2006 May 29;580(13):3192-200. doi: 10.1016/j.febslet.2006.04.076. Epub 2006 May 4. FEBS Lett. 2006. PMID: 16684538 Free PMC article.
-
Combating the challenges of COVID-19 pandemic: Insights into molecular mechanisms, immune responses and therapeutics against SARS-CoV-2.Oxf Open Immunol. 2023 Jan 10;4(1):iqad001. doi: 10.1093/oxfimm/iqad001. eCollection 2023. Oxf Open Immunol. 2023. PMID: 37051070 Free PMC article. Review.
-
Structural Characterization of SARS-CoV-2: Where We Are, and Where We Need to Be.Front Mol Biosci. 2020 Dec 17;7:605236. doi: 10.3389/fmolb.2020.605236. eCollection 2020. Front Mol Biosci. 2020. PMID: 33392262 Free PMC article. Review.
References
-
- Corse E., Machamer C.E. Infectious bronchitis virus envelope protein targeting: implications for virus assembly. Adv Exp. Med. Biol. 2001;494:571–576. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

