The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex
- PMID: 12885899
- PMCID: PMC167208
- DOI: 10.1128/jvi.77.16.8801-8811.2003
The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex
Abstract
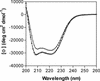
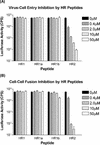
Coronavirus entry is mediated by the viral spike (S) glycoprotein. The 180-kDa oligomeric S protein of the murine coronavirus mouse hepatitis virus strain A59 is posttranslationally cleaved into an S1 receptor binding unit and an S2 membrane fusion unit. The latter is thought to contain an internal fusion peptide and has two 4,3 hydrophobic (heptad) repeat regions designated HR1 and HR2. HR2 is located close to the membrane anchor, and HR1 is some 170 amino acids (aa) upstream of it. Heptad repeat (HR) regions are found in fusion proteins of many different viruses and form an important characteristic of class I viral fusion proteins. We investigated the role of these regions in coronavirus membrane fusion. Peptides HR1 (96 aa) and HR2 (39 aa), corresponding to the HR1 and HR2 regions, were produced in Escherichia coli. When mixed together, the two peptides were found to assemble into an extremely stable oligomeric complex. Both on their own and within the complex, the peptides were highly alpha helical. Electron microscopic analysis of the complex revealed a rod-like structure approximately 14.5 nm in length. Limited proteolysis in combination with mass spectrometry indicated that HR1 and HR2 occur in the complex in an antiparallel fashion. In the native protein, such a conformation would bring the proposed fusion peptide, located in the N-terminal domain of HR1, and the transmembrane anchor into close proximity. Using biological assays, the HR2 peptide was shown to be a potent inhibitor of virus entry into the cell, as well as of cell-cell fusion. Both biochemical and functional data show that the coronavirus spike protein is a class I viral fusion protein.
Figures






Similar articles
-
Coronavirus escape from heptad repeat 2 (HR2)-derived peptide entry inhibition as a result of mutations in the HR1 domain of the spike fusion protein.J Virol. 2008 Mar;82(5):2580-5. doi: 10.1128/JVI.02287-07. Epub 2007 Dec 12. J Virol. 2008. PMID: 18077706 Free PMC article.
-
Interaction between heptad repeat 1 and 2 regions in spike protein of SARS-associated coronavirus: implications for virus fusogenic mechanism and identification of fusion inhibitors.Lancet. 2004 Mar 20;363(9413):938-47. doi: 10.1016/S0140-6736(04)15788-7. Lancet. 2004. PMID: 15043961 Free PMC article.
-
Structural basis for coronavirus-mediated membrane fusion. Crystal structure of mouse hepatitis virus spike protein fusion core.J Biol Chem. 2004 Jul 16;279(29):30514-22. doi: 10.1074/jbc.M403760200. Epub 2004 Apr 27. J Biol Chem. 2004. PMID: 15123674 Free PMC article.
-
Site directed mutagenesis of the murine coronavirus spike protein. Effects on fusion.Adv Exp Med Biol. 1995;380:283-6. doi: 10.1007/978-1-4615-1899-0_45. Adv Exp Med Biol. 1995. PMID: 8830493 Review.
-
[Cell entry mechanisms of coronaviruses].Uirusu. 2009 Dec;59(2):215-22. doi: 10.2222/jsv.59.215. Uirusu. 2009. PMID: 20218330 Review. Japanese.
Cited by
-
[Biology and pathology of coronaviruses].Pathologe. 2021 Mar;42(2):149-154. doi: 10.1007/s00292-021-00923-y. Epub 2021 Mar 1. Pathologe. 2021. PMID: 33646361 Free PMC article. Review. German.
-
Structures and dynamics of the novel S1/S2 protease cleavage site loop of the SARS-CoV-2 spike glycoprotein.J Struct Biol X. 2020;4:100038. doi: 10.1016/j.yjsbx.2020.100038. Epub 2020 Oct 5. J Struct Biol X. 2020. PMID: 33043289 Free PMC article.
-
An in silico approach for identification of novel inhibitors as potential therapeutics targeting COVID-19 main protease.J Biomol Struct Dyn. 2021 Aug;39(12):4304-4315. doi: 10.1080/07391102.2020.1776158. Epub 2020 Jun 16. J Biomol Struct Dyn. 2021. PMID: 32544024 Free PMC article.
-
Immunoinformatics characterization of SARS-CoV-2 spike glycoprotein for prioritization of epitope based multivalent peptide vaccine.J Mol Liq. 2020 Sep 15;314:113612. doi: 10.1016/j.molliq.2020.113612. Epub 2020 Jun 17. J Mol Liq. 2020. PMID: 32834259 Free PMC article.
-
In Silico Identification of Potential Inhibitors of ADP-Ribose Phosphatase of SARS-CoV-2 nsP3 by Combining E-Pharmacophore- and Receptor-Based Virtual Screening of Database.ChemistrySelect. 2020 Aug 14;5(30):9388-9398. doi: 10.1002/slct.202001419. Epub 2020 Aug 11. ChemistrySelect. 2020. PMID: 32835090 Free PMC article.
References
-
- Baker, K. A., R. E. Dutch, R. A. Lamb, and T. S. Jardetzky. 1999. Structural basis for paramyxovirus-mediated membrane fusion. Mol. Cell 3:309-319. - PubMed
-
- Buchholz, U. J., S. Finke, and K. K. Conzelmann. 1999. Generation of bovine respiratory syncytial virus (BRSV) from cDNA: BRSV NS2 is not essential for virus replication in tissue culture, and the human RSV leader region acts as a functional BRSV genome promoter. J. Virol. 73:251-259. - PMC - PubMed
-
- Bullough, P. A., F. M. Hughson, J. J. Skehel, and D. C. Wiley. 1994. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature 371:37-43. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

