Dynamic, yet structured: The cell membrane three decades after the Singer-Nicolson model
- PMID: 12832616
- PMCID: PMC166180
- DOI: 10.1073/pnas.1332550100
Dynamic, yet structured: The cell membrane three decades after the Singer-Nicolson model
Abstract
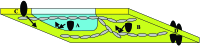
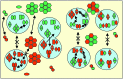
The fluid mosaic membrane model proved to be a very useful hypothesis in explaining many, but certainly not all, phenomena taking place in biological membranes. New experimental data show that the compartmentalization of membrane components can be as important for effective signal transduction as is the fluidity of the membrane. In this work, we pay tribute to the Singer-Nicolson model, which is near its 30th anniversary, honoring its basic features, "mosaicism" and "diffusion," which predict the interspersion of proteins and lipids and their ability to undergo dynamic rearrangement via Brownian motion. At the same time, modifications based on quantitative data are proposed, highlighting the often genetically predestined, yet flexible, multilevel structure implementing a vast complexity of cellular functions. This new "dynamically structured mosaic model" bears the following characteristics: emphasis is shifted from fluidity to mosaicism, which, in our interpretation, means nonrandom codistribution patterns of specific kinds of membrane proteins forming small-scale clusters at the molecular level and large-scale clusters (groups of clusters, islands) at the submicrometer level. The cohesive forces, which maintain these assemblies as principal elements of the membranes, originate from within a microdomain structure, where lipid-lipid, protein-protein, and protein-lipid interactions, as well as sub- and supramembrane (cytoskeletal, extracellular matrix, other cell) effectors, many of them genetically predestined, play equally important roles. The concept of fluidity in the original model now is interpreted as permissiveness of the architecture to continuous, dynamic restructuring of the molecular- and higher-level clusters according to the needs of the cell and as evoked by the environment.
Figures
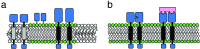
Similar articles
-
The Fluid-Mosaic Model of Membrane Structure: still relevant to understanding the structure, function and dynamics of biological membranes after more than 40 years.Biochim Biophys Acta. 2014 Jun;1838(6):1451-66. doi: 10.1016/j.bbamem.2013.10.019. Epub 2013 Nov 1. Biochim Biophys Acta. 2014. PMID: 24189436 Review.
-
Macromolecular crowding: chemistry and physics meet biology (Ascona, Switzerland, 10-14 June 2012).Phys Biol. 2013 Aug;10(4):040301. doi: 10.1088/1478-3975/10/4/040301. Epub 2013 Aug 2. Phys Biol. 2013. PMID: 23912807
-
The Fluid-Mosaic model of cell membranes: A brief introduction, historical features, some general principles, and its adaptation to current information.Biochim Biophys Acta Biomembr. 2023 Apr;1865(4):184135. doi: 10.1016/j.bbamem.2023.184135. Epub 2023 Feb 5. Biochim Biophys Acta Biomembr. 2023. PMID: 36746313 Review.
-
Is a fluid-mosaic model of biological membranes fully relevant? Studies on lipid organization in model and biological membranes.Cell Mol Biol Lett. 2003;8(1):147-59. Cell Mol Biol Lett. 2003. PMID: 12655369
-
Dynamics in the plasma membrane: how to combine fluidity and order.EMBO J. 2006 Aug 9;25(15):3446-57. doi: 10.1038/sj.emboj.7601204. Epub 2006 Jun 22. EMBO J. 2006. PMID: 16900097 Free PMC article. Review.
Cited by
-
Inferring diffusion in single live cells at the single-molecule level.Philos Trans R Soc Lond B Biol Sci. 2012 Dec 24;368(1611):20120029. doi: 10.1098/rstb.2012.0029. Print 2013 Feb 5. Philos Trans R Soc Lond B Biol Sci. 2012. PMID: 23267182 Free PMC article. Review.
-
Alteration of interleaflet coupling due to compounds displaying rapid translocation in lipid membranes.Sci Rep. 2016 Sep 6;6:32934. doi: 10.1038/srep32934. Sci Rep. 2016. PMID: 27596355 Free PMC article.
-
The Lipidome Fingerprint of Longevity.Molecules. 2020 Sep 22;25(18):4343. doi: 10.3390/molecules25184343. Molecules. 2020. PMID: 32971886 Free PMC article. Review.
-
Drug resistance in breast cancer cells: biophysical characterization of and doxorubicin interactions with membrane lipids.Mol Pharm. 2010 Dec 6;7(6):2334-48. doi: 10.1021/mp100308n. Epub 2010 Nov 11. Mol Pharm. 2010. PMID: 20958074 Free PMC article.
-
Atomic-level description of protein-lipid interactions using an accelerated membrane model.Biochim Biophys Acta. 2016 Jul;1858(7 Pt B):1573-83. doi: 10.1016/j.bbamem.2016.02.027. Epub 2016 Mar 2. Biochim Biophys Acta. 2016. PMID: 26940626 Free PMC article. Review.
References
-
- Singer, S. J. & Nicolson, G. L. (1972) Science 175 720-731. - PubMed
-
- Jacobson, K., Sheets, E. D. & Simson, R. (1995) Science 268 1441-1442. - PubMed
-
- Sheets, E. D., Simson, R. & Jacobson, K. (1995) Curr. Opin. Cell Biol. 7 707-714. - PubMed
-
- Damjanovich, S., Gáspár, R., Jr., & Pieri, C. (1997) Q. Rev. Biophys. 30 67-106. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

