Overexpression of the cochaperone CHIP enhances Hsp70-dependent folding activity in mammalian cells
- PMID: 12832480
- PMCID: PMC162225
- DOI: 10.1128/MCB.23.14.4948-4958.2003
Overexpression of the cochaperone CHIP enhances Hsp70-dependent folding activity in mammalian cells
Abstract
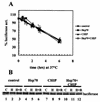
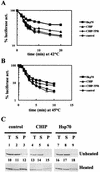
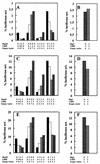
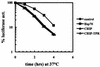
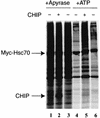
CHIP is a cochaperone of Hsp70 that inhibits Hsp70-dependent refolding in vitro. However, the effect of altered expression of CHIP on the fate of unfolded proteins in mammalian cells has not been determined. Surprisingly, we found that overexpression of CHIP in fibroblasts increased the refolding of proteins after thermal denaturation. This effect was insensitive to geldanamycin, an Hsp90 inhibitor, and required the tetratricopeptide repeat motifs but not the U-box domain of CHIP. Inhibition of Hsp70 chaperone activity abolished the effects of CHIP on protein folding, indicating that the CHIP-mediated events were Hsp70 dependent. Hsp40 competitively inhibited the CHIP-dependent refolding, which is consistent with in vitro data indicating that these cofactors act on Hsp70 in the ATP-bound state and have opposing effects on Hsp70 ATPase activity. Consistent with these observations, CHIP overexpression did not alter protein folding in the setting of ATP depletion, when Hsp70 is in the ADP-bound state. Concomitant with its effects on refolding heat-denatured substrates, CHIP increased the fraction of nascent chains coimmunoprecipitating with Hsc70, but only when sufficient ATP was present to allow Hsp70 to cycle rapidly. Our data suggest that, consistent with in vitro studies, CHIP attenuates the Hsp70 cycle in living cells. The impact of this effect on the fate of unfolded proteins in cells, however, is different from what might be expected from the in vitro data. Rather than resulting in inhibited refolding, CHIP increases the folding capacity of Hsp70 in eukaryotic cells.
Figures


Similar articles
-
Cofactor Tpr2 combines two TPR domains and a J domain to regulate the Hsp70/Hsp90 chaperone system.EMBO J. 2003 Jul 15;22(14):3613-23. doi: 10.1093/emboj/cdg362. EMBO J. 2003. PMID: 12853476 Free PMC article.
-
Both the N- and C-terminal chaperone sites of Hsp90 participate in protein refolding.Eur J Biochem. 2001 Apr;268(8):2520-4. doi: 10.1046/j.1432-1327.2001.02145.x. Eur J Biochem. 2001. PMID: 11298772
-
Identification of CHIP, a novel tetratricopeptide repeat-containing protein that interacts with heat shock proteins and negatively regulates chaperone functions.Mol Cell Biol. 1999 Jun;19(6):4535-45. doi: 10.1128/MCB.19.6.4535. Mol Cell Biol. 1999. PMID: 10330192 Free PMC article.
-
The Hsp70-Hsp90 Chaperone Cascade in Protein Folding.Trends Cell Biol. 2019 Feb;29(2):164-177. doi: 10.1016/j.tcb.2018.10.004. Epub 2018 Nov 28. Trends Cell Biol. 2019. PMID: 30502916 Review.
-
Regulation of signaling protein function and trafficking by the hsp90/hsp70-based chaperone machinery.Exp Biol Med (Maywood). 2003 Feb;228(2):111-33. doi: 10.1177/153537020322800201. Exp Biol Med (Maywood). 2003. PMID: 12563018 Review.
Cited by
-
Opposing actions of heat shock protein 90 and 70 regulate nicotinamide adenine dinucleotide phosphate oxidase stability and reactive oxygen species production.Arterioscler Thromb Vasc Biol. 2012 Dec;32(12):2989-99. doi: 10.1161/ATVBAHA.112.300361. Epub 2012 Sep 27. Arterioscler Thromb Vasc Biol. 2012. PMID: 23023377 Free PMC article.
-
Molecular chaperones in Parkinson's disease--present and future.J Parkinsons Dis. 2011;1(4):299-320. J Parkinsons Dis. 2011. PMID: 22279517 Free PMC article. Review.
-
Molecular chaperones as rational drug targets for Parkinson's disease therapeutics.CNS Neurol Disord Drug Targets. 2010 Dec;9(6):741-53. doi: 10.2174/187152710793237386. CNS Neurol Disord Drug Targets. 2010. PMID: 20942788 Free PMC article. Review.
-
Most mutations that cause spinocerebellar ataxia autosomal recessive type 16 (SCAR16) destabilize the protein quality-control E3 ligase CHIP.J Biol Chem. 2018 Feb 23;293(8):2735-2743. doi: 10.1074/jbc.RA117.000477. Epub 2018 Jan 9. J Biol Chem. 2018. PMID: 29317501 Free PMC article.
-
Control of AIF-mediated cell death by antagonistic functions of CHIP ubiquitin E3 ligase and USP2 deubiquitinating enzyme.Cell Death Differ. 2011 Aug;18(8):1326-36. doi: 10.1038/cdd.2011.3. Epub 2011 Feb 4. Cell Death Differ. 2011. PMID: 21293491 Free PMC article.
References
-
- Aravind, L., and E. V. Koonin. 2000. The U box is a modified RING finger — a common domain in ubiquitination. Curr. Biol. 10:R132-R134. - PubMed
-
- Ballinger, C. A., P. Connell, Y. Wu, Z. Hu, L. J. Thompson, L. Y. Yin, and C. Patterson. 1999. Identification of CHIP, a novel tetratricopeptide repeat-containing protein that interacts with heat shock proteins and negatively regulates chaperone functions. Mol. Cell. Biol. 19:4535-4545. - PMC - PubMed
-
- Beckmann, R. P., L. A. Mizzen, and W. J. Welch. 1990. Interaction of Hsp 70 with newly synthesized proteins: implications for protein folding and assembly. Science 248:850-854. - PubMed
-
- Bukau, B., and A. L. Horwich. 1998. The Hsp70 and Hsp60 chaperone machines. Cell 92:351-366. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
