Slx1-Slx4 is a second structure-specific endonuclease functionally redundant with Sgs1-Top3
- PMID: 12832395
- PMCID: PMC196184
- DOI: 10.1101/gad.1105203
Slx1-Slx4 is a second structure-specific endonuclease functionally redundant with Sgs1-Top3
Abstract
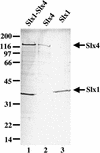
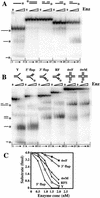
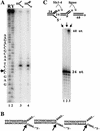
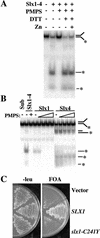
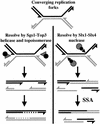
The RecQ DNA helicases human BLM and yeast Sgs1 interact with DNA topoisomerase III and are thought to act on stalled replication forks to maintain genome stability. To gain insight into this mechanism, we previously identified SLX1 and SLX4 as genes that are required for viability and for completion of rDNA replication in the absence of SGS1-TOP3. Here we show that SLX1 and SLX4 encode a heteromeric structure-specific endonuclease. The Slx1-Slx4 nuclease is active on branched DNA substrates, particularly simple-Y, 5'-flap, or replication fork structures. It cleaves the strand bearing the 5' nonhomologous arm at the branch junction and generates ligatable nicked products from 5'-flap or replication fork substrates. Slx1 is the founding member of a family of proteins with a predicted URI nuclease domain and PHD-type zinc finger. This subunit displays weak structure-specific endonuclease activity on its own, is stimulated 500-fold by Slx4, and requires the PHD finger for activity in vitro and in vivo. Both subunits are required in vivo for resistance to DNA damage by methylmethane sulfonate (MMS). We propose that Sgs1-Top3 acts at the termination of rDNA replication to decatenate stalled forks, and, in its absence, Slx1-Slx4 cleaves these stalled forks.
Figures


Similar articles
-
Functional overlap between Sgs1-Top3 and the Mms4-Mus81 endonuclease.Genes Dev. 2001 Oct 15;15(20):2730-40. doi: 10.1101/gad.932201. Genes Dev. 2001. PMID: 11641278 Free PMC article.
-
Yeast Rmi1/Nce4 controls genome stability as a subunit of the Sgs1-Top3 complex.Mol Cell Biol. 2005 Jun;25(11):4476-87. doi: 10.1128/MCB.25.11.4476-4487.2005. Mol Cell Biol. 2005. PMID: 15899853 Free PMC article.
-
Requirement for three novel protein complexes in the absence of the Sgs1 DNA helicase in Saccharomyces cerevisiae.Genetics. 2001 Jan;157(1):103-18. doi: 10.1093/genetics/157.1.103. Genetics. 2001. PMID: 11139495 Free PMC article.
-
[Functional analysis of yeast homologue gene associated with human DNA helicase causative syndromes].Kokuritsu Iyakuhin Shokuhin Eisei Kenkyusho Hokoku. 2002;(120):53-74. Kokuritsu Iyakuhin Shokuhin Eisei Kenkyusho Hokoku. 2002. PMID: 12638184 Review. Japanese.
-
Yeast as a model system to study RecQ helicase function.DNA Repair (Amst). 2010 Mar 2;9(3):303-14. doi: 10.1016/j.dnarep.2009.12.007. Epub 2010 Jan 13. DNA Repair (Amst). 2010. PMID: 20071248 Review.
Cited by
-
Novel insights into RAD51 activity and regulation during homologous recombination and DNA replication.Biochem Cell Biol. 2016 Oct;94(5):407-418. doi: 10.1139/bcb-2016-0012. Epub 2016 Mar 31. Biochem Cell Biol. 2016. PMID: 27224545 Free PMC article. Review.
-
GEN1/Yen1 and the SLX4 complex: Solutions to the problem of Holliday junction resolution.Genes Dev. 2010 Mar 15;24(6):521-36. doi: 10.1101/gad.1903510. Epub 2010 Mar 4. Genes Dev. 2010. PMID: 20203129 Free PMC article. Review.
-
A tale of tails: insights into the coordination of 3' end processing during homologous recombination.Bioessays. 2009 Mar;31(3):315-21. doi: 10.1002/bies.200800195. Bioessays. 2009. PMID: 19260026 Free PMC article. Review.
-
The many lives of type IA topoisomerases.J Biol Chem. 2020 May 15;295(20):7138-7153. doi: 10.1074/jbc.REV120.008286. Epub 2020 Apr 10. J Biol Chem. 2020. PMID: 32277049 Free PMC article. Review.
-
Ultrafine anaphase bridges, broken DNA and illegitimate recombination induced by a replication fork barrier.Nucleic Acids Res. 2011 Aug;39(15):6568-84. doi: 10.1093/nar/gkr340. Epub 2011 May 16. Nucleic Acids Res. 2011. PMID: 21576223 Free PMC article.
References
-
- Aasland R., Gibson, T.J., and Stewart, A.F. 1995. The PHD finger: Implications for chromatin-mediated transcriptional regulation. Trends Biochem. Sci. 20: 56–59. - PubMed
-
- Aravind L. and Koonin, E.V. 2000. SAP—A putative DNA-binding motif involved in chromosomal organization. Trends Biochem. Sci. 25: 112–114. - PubMed
-
- Bochkareva E., Frappier, L., Edwards, A.M., and Bochkarev, A. 1998. The RPA32 subunit of human replication protein A contains a single-stranded DNA-binding domain. J. Biol. Chem. 273: 3932–3936. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
