Pfold: RNA secondary structure prediction using stochastic context-free grammars
- PMID: 12824339
- PMCID: PMC169020
- DOI: 10.1093/nar/gkg614
Pfold: RNA secondary structure prediction using stochastic context-free grammars
Abstract
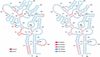
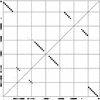
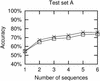
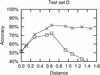
RNA secondary structures are important in many biological processes and efficient structure prediction can give vital directions for experimental investigations. Many available programs for RNA secondary structure prediction only use a single sequence at a time. This may be sufficient in some applications, but often it is possible to obtain related RNA sequences with conserved secondary structure. These should be included in structural analyses to give improved results. This work presents a practical way of predicting RNA secondary structure that is especially useful when related sequences can be obtained. The method improves a previous algorithm based on an explicit evolutionary model and a probabilistic model of structures. Predictions can be done on a web server at http://www.daimi.au.dk/~compbio/pfold.
Figures
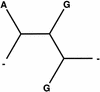

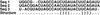


Similar articles
-
Pair stochastic tree adjoining grammars for aligning and predicting pseudoknot RNA structures.Proc IEEE Comput Syst Bioinform Conf. 2004:290-9. Proc IEEE Comput Syst Bioinform Conf. 2004. PMID: 16448022
-
Multithreaded comparative RNA secondary structure prediction using stochastic context-free grammars.BMC Bioinformatics. 2011 Apr 18;12:103. doi: 10.1186/1471-2105-12-103. BMC Bioinformatics. 2011. PMID: 21501497 Free PMC article.
-
SCFGs in RNA secondary structure prediction RNA secondary structure prediction: a hands-on approach.Methods Mol Biol. 2014;1097:143-62. doi: 10.1007/978-1-62703-709-9_8. Methods Mol Biol. 2014. PMID: 24639159
-
Energy-based RNA consensus secondary structure prediction in multiple sequence alignments.Methods Mol Biol. 2014;1097:125-41. doi: 10.1007/978-1-62703-709-9_7. Methods Mol Biol. 2014. PMID: 24639158 Review.
-
Introduction to stochastic context free grammars.Methods Mol Biol. 2014;1097:85-106. doi: 10.1007/978-1-62703-709-9_5. Methods Mol Biol. 2014. PMID: 24639156 Review.
Cited by
-
Thirteen dubious ways to detect conserved structural RNAs.IUBMB Life. 2023 Jun;75(6):471-492. doi: 10.1002/iub.2694. Epub 2022 Dec 10. IUBMB Life. 2023. PMID: 36495545 Free PMC article.
-
RNA folding using quantum computers.PLoS Comput Biol. 2022 Apr 11;18(4):e1010032. doi: 10.1371/journal.pcbi.1010032. eCollection 2022 Apr. PLoS Comput Biol. 2022. PMID: 35404931 Free PMC article.
-
What's Wrong in a Jump? Prediction and Validation of Splice Site Variants.Methods Protoc. 2021 Sep 5;4(3):62. doi: 10.3390/mps4030062. Methods Protoc. 2021. PMID: 34564308 Free PMC article. Review.
-
The effects of structure on pre-mRNA processing and stability.Methods. 2017 Aug 1;125:36-44. doi: 10.1016/j.ymeth.2017.06.001. Epub 2017 Jun 6. Methods. 2017. PMID: 28595983 Free PMC article. Review.
-
Advances and opportunities in RNA structure experimental determination and computational modeling.Nat Methods. 2022 Oct;19(10):1193-1207. doi: 10.1038/s41592-022-01623-y. Epub 2022 Oct 6. Nat Methods. 2022. PMID: 36203019 Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

