MAX4 and RMS1 are orthologous dioxygenase-like genes that regulate shoot branching in Arabidopsis and pea
- PMID: 12815068
- PMCID: PMC196077
- DOI: 10.1101/gad.256603
MAX4 and RMS1 are orthologous dioxygenase-like genes that regulate shoot branching in Arabidopsis and pea
Abstract
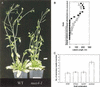
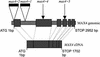
Shoot branching is inhibited by auxin transported down the stem from the shoot apex. Auxin does not accumulate in inhibited buds and so must act indirectly. We show that mutations in the MAX4 gene of Arabidopsis result in increased and auxin-resistant bud growth. Increased branching in max4 shoots is restored to wild type by grafting to wild-type rootstocks, suggesting that MAX4 is required to produce a mobile branch-inhibiting signal, acting downstream of auxin. A similar role has been proposed for the pea gene, RMS1. Accordingly, MAX4 and RMS1 were found to encode orthologous, auxin-inducible members of the polyene dioxygenase family.
Figures

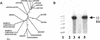

Similar articles
-
Hormonally controlled expression of the Arabidopsis MAX4 shoot branching regulatory gene.Plant J. 2005 Nov;44(4):569-80. doi: 10.1111/j.1365-313X.2005.02548.x. Plant J. 2005. PMID: 16262707
-
The Arabidopsis MAX pathway controls shoot branching by regulating auxin transport.Curr Biol. 2006 Mar 21;16(6):553-63. doi: 10.1016/j.cub.2006.01.058. Curr Biol. 2006. PMID: 16546078
-
The branching gene RAMOSUS1 mediates interactions among two novel signals and auxin in pea.Plant Cell. 2005 Feb;17(2):464-74. doi: 10.1105/tpc.104.026716. Epub 2005 Jan 19. Plant Cell. 2005. PMID: 15659639 Free PMC article.
-
The fall and rise of apical dominance.Curr Opin Genet Dev. 2005 Aug;15(4):468-71. doi: 10.1016/j.gde.2005.06.010. Curr Opin Genet Dev. 2005. PMID: 15964756 Review.
-
Regulation of shoot and root development through mutual signaling.Mol Plant. 2012 Sep;5(5):974-83. doi: 10.1093/mp/sss047. Epub 2012 May 24. Mol Plant. 2012. PMID: 22628542 Review.
Cited by
-
Functions of exogenous strigolactone application and strigolactone biosynthesis genes GhMAX3/GhMAX4b in response to drought tolerance in cotton (Gossypium hirsutum L.).BMC Plant Biol. 2024 Oct 26;24(1):1008. doi: 10.1186/s12870-024-05726-w. BMC Plant Biol. 2024. PMID: 39455926 Free PMC article.
-
Strigolactone: An Emerging Growth Regulator for Developing Resilience in Plants.Plants (Basel). 2022 Oct 3;11(19):2604. doi: 10.3390/plants11192604. Plants (Basel). 2022. PMID: 36235470 Free PMC article. Review.
-
Comparative Transcriptomic Analysis of Two Actinorhizal Plants and the Legume Medicago truncatula Supports the Homology of Root Nodule Symbioses and Is Congruent With a Two-Step Process of Evolution in the Nitrogen-Fixing Clade of Angiosperms.Front Plant Sci. 2018 Oct 8;9:1256. doi: 10.3389/fpls.2018.01256. eCollection 2018. Front Plant Sci. 2018. PMID: 30349546 Free PMC article.
-
Identification of cis-elements that regulate gene expression during initiation of axillary bud outgrowth in Arabidopsis.Plant Physiol. 2005 Jun;138(2):757-66. doi: 10.1104/pp.104.057984. Epub 2005 May 20. Plant Physiol. 2005. PMID: 15908603 Free PMC article.
-
Strigolactone regulation of shoot branching in chrysanthemum (Dendranthema grandiflorum).J Exp Bot. 2010 Jun;61(11):3069-78. doi: 10.1093/jxb/erq133. Epub 2010 May 17. J Exp Bot. 2010. PMID: 20478970 Free PMC article.
References
-
- Altschul S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403–410. - PubMed
-
- Bangerth F. 1994. Response of cytokinin concentration in the xylem exudate of bean (Phaseolus vulgaris L.) plants to decapitation and auxin treatment, and relationship to apical dominance. Planta 194: 439–442.
-
- Beveridge C.A. 2000. Long-distance signaling and a mutational analysis of branching in pea. Plant Growth Regul. 32: 193–203.
-
- Beveridge C.A., Symons, G.M., Murfet, I.C., Ross, J.J., and Rameau, C. 1997. The rms1 mutant of pea has elevated indole-3-acetic acid levels and reduced root-sap zeatin riboside content but increased branching controlled by graft-transmissible signals. Plant Physiol. 115: 1251–1258.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
