H1 linker histones are essential for mouse development and affect nucleosome spacing in vivo
- PMID: 12808097
- PMCID: PMC164858
- DOI: 10.1128/MCB.23.13.4559-4572.2003
H1 linker histones are essential for mouse development and affect nucleosome spacing in vivo
Abstract
Most eukaryotic cells contain nearly equimolar amounts of nucleosomes and H1 linker histones. Despite their abundance and the potential functional specialization of H1 subtypes in multicellular organisms, gene inactivation studies have failed to reveal essential functions for linker histones in vivo. Moreover, in vitro studies suggest that H1 subtypes may not be absolutely required for assembly of chromosomes or nuclei. By sequentially inactivating the genes for three mouse H1 subtypes (H1c, H1d, and H1e), we showed that linker histones are essential for mammalian development. Embryos lacking the three H1 subtypes die by mid-gestation with a broad range of defects. Triple-H1-null embryos have about 50% of the normal ratio of H1 to nucleosomes. Mice null for five of these six H1 alleles are viable but are underrepresented in litters and are much smaller than their littermates. Marked reductions in H1 content were found in certain tissues of these mice and in another compound H1 mutant. These results demonstrate that the total amount of H1 is crucial for proper embryonic development. Extensive reduction of H1 in certain tissues did not lead to changes in nuclear size, but it did result in global shortening of the spacing between nucleosomes.
Figures
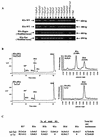
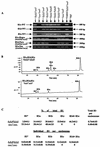
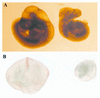




Similar articles
-
Individual somatic H1 subtypes are dispensable for mouse development even in mice lacking the H1(0) replacement subtype.Mol Cell Biol. 2001 Dec;21(23):7933-43. doi: 10.1128/MCB.21.23.7933-7943.2001. Mol Cell Biol. 2001. PMID: 11689686 Free PMC article.
-
Reduction of Hox gene expression by histone H1 depletion.PLoS One. 2012;7(6):e38829. doi: 10.1371/journal.pone.0038829. Epub 2012 Jun 11. PLoS One. 2012. PMID: 22701719 Free PMC article.
-
Novel nucleosomal particles containing core histones and linker DNA but no histone H1.Nucleic Acids Res. 2016 Jan 29;44(2):573-81. doi: 10.1093/nar/gkv943. Epub 2015 Sep 22. Nucleic Acids Res. 2016. PMID: 26400169 Free PMC article.
-
Role of linker histone in chromatin structure and function: H1 stoichiometry and nucleosome repeat length.Chromosome Res. 2006;14(1):17-25. doi: 10.1007/s10577-005-1024-3. Chromosome Res. 2006. PMID: 16506093 Review.
-
The H1 linker histones: multifunctional proteins beyond the nucleosomal core particle.EMBO Rep. 2015 Nov;16(11):1439-53. doi: 10.15252/embr.201540749. Epub 2015 Oct 15. EMBO Rep. 2015. PMID: 26474902 Free PMC article. Review.
Cited by
-
Comparative analysis of linker histone H1, MeCP2, and HMGD1 on nucleosome stability and target site accessibility.Sci Rep. 2016 Sep 14;6:33186. doi: 10.1038/srep33186. Sci Rep. 2016. PMID: 27624769 Free PMC article.
-
Histone H1 Limits DNA Methylation in Neurospora crassa.G3 (Bethesda). 2016 Jul 7;6(7):1879-89. doi: 10.1534/g3.116.028324. G3 (Bethesda). 2016. PMID: 27172195 Free PMC article.
-
The Epstein-Barr virus nuclear antigen-1 reprograms transcription by mimicry of high mobility group A proteins.Nucleic Acids Res. 2013 Mar 1;41(5):2950-62. doi: 10.1093/nar/gkt032. Epub 2013 Jan 28. Nucleic Acids Res. 2013. PMID: 23358825 Free PMC article.
-
Genetic and epigenetic mechanisms of gene regulation during lens development.Prog Retin Eye Res. 2007 Nov;26(6):555-97. doi: 10.1016/j.preteyeres.2007.07.002. Epub 2007 Jul 28. Prog Retin Eye Res. 2007. PMID: 17905638 Free PMC article. Review.
-
Linker histone phosphorylation regulates global timing of replication origin firing.J Biol Chem. 2009 Jan 30;284(5):2823-2829. doi: 10.1074/jbc.M805617200. Epub 2008 Nov 17. J Biol Chem. 2009. PMID: 19015270 Free PMC article.
References
-
- Adenot, P. G., E. Campion, E. Legouy, C. D. Allis, S. Dimitrov, J. Renard, and E. M. Thompson. 2000. Somatic linker histone H1 is present throughout mouse embryogenesis and is not replaced by variant H1 degrees. J. Cell Sci.113:2897-2907. - PubMed
-
- Annunziato, A. T., and R. L. Seale. 1983. Chromatin replication, reconstitution and assembly. Mol. Cell. Biochem. 55:99-112. - PubMed
-
- Blank, T. A., and P. B. Becker. 1995. Electrostatic mechanism of nucleosome spacing. J. Mol. Biol. 252:305-313. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
