Mechanisms of activation, inhibition and specificity: crystal structures of the NMDA receptor NR1 ligand-binding core
- PMID: 12805203
- PMCID: PMC162155
- DOI: 10.1093/emboj/cdg303
Mechanisms of activation, inhibition and specificity: crystal structures of the NMDA receptor NR1 ligand-binding core
Abstract
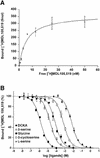
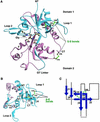
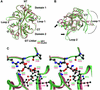
Excitatory neurotransmission mediated by the N-methyl-D-aspartate subtype of ionotropic glutamate receptors is fundamental to the development and function of the mammalian central nervous system. NMDA receptors require both glycine and glutamate for activation with NR1 and NR2 forming glycine and glutamate sites, respectively. Mechanisms to describe agonist and antagonist binding, and activation and desensitization of NMDA receptors have been hampered by the lack of high-resolution structures. Here, we describe the cocrystal structures of the NR1 S1S2 ligand-binding core with the agonists glycine and D-serine (DS), the partial agonist D-cycloserine (DCS) and the antagonist 5,7-dichlorokynurenic acid (DCKA). The cleft of the S1S2 'clamshell' is open in the presence of the antagonist DCKA and closed in the glycine, DS and DCS complexes. In addition, the NR1 S1S2 structure reveals the fold and interactions of loop 1, a cysteine-rich region implicated in intersubunit allostery.
Figures

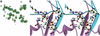

Similar articles
-
Model structures of the N-methyl-D-aspartate receptor subunit NR1 explain the molecular recognition of agonist and antagonist ligands.J Struct Biol. 2004 Mar;145(3):205-15. doi: 10.1016/j.jsb.2003.10.016. J Struct Biol. 2004. PMID: 14960371
-
Evolutionary trace analysis of ionotropic glutamate receptor sequences and modeling the interactions of agonists with different NMDA receptor subunits.J Mol Model. 2004 Dec;10(5-6):305-16. doi: 10.1007/s00894-004-0196-7. Epub 2004 Oct 22. J Mol Model. 2004. PMID: 15597199
-
Structural consequences of D481N/K483Q mutation at glycine binding site of NMDA ionotropic glutamate receptors: a molecular dynamics study.J Biomol Struct Dyn. 2005 Feb;22(4):399-410. doi: 10.1080/07391102.2005.10507012. J Biomol Struct Dyn. 2005. PMID: 15588104
-
Influence of the NR3A subunit on NMDA receptor functions.Prog Neurobiol. 2010 May;91(1):23-37. doi: 10.1016/j.pneurobio.2010.01.004. Epub 2010 Jan 25. Prog Neurobiol. 2010. PMID: 20097255 Free PMC article. Review.
-
Shuffling the deck anew: how NR3 tweaks NMDA receptor function.Mol Neurobiol. 2008 Aug;38(1):16-26. doi: 10.1007/s12035-008-8029-9. Epub 2008 Jul 25. Mol Neurobiol. 2008. PMID: 18654865 Review.
Cited by
-
Modes of glutamate receptor gating.J Physiol. 2012 Jan 1;590(1):73-91. doi: 10.1113/jphysiol.2011.223750. Epub 2011 Nov 21. J Physiol. 2012. PMID: 22106181 Free PMC article. Review.
-
Emerging structural insights into the function of ionotropic glutamate receptors.Trends Biochem Sci. 2015 Jun;40(6):328-37. doi: 10.1016/j.tibs.2015.04.002. Epub 2015 May 1. Trends Biochem Sci. 2015. PMID: 25941168 Free PMC article. Review.
-
Radical pairs may play a role in xenon-induced general anesthesia.Sci Rep. 2021 Mar 18;11(1):6287. doi: 10.1038/s41598-021-85673-w. Sci Rep. 2021. PMID: 33737599 Free PMC article.
-
Structural mechanisms of activation and desensitization in neurotransmitter-gated ion channels.Nat Struct Mol Biol. 2016 Jun 7;23(6):494-502. doi: 10.1038/nsmb.3214. Nat Struct Mol Biol. 2016. PMID: 27273633 Review.
-
Discovery of (R)-2-amino-3-triazolpropanoic acid derivatives as NMDA receptor glycine site agonists with GluN2 subunit-specific activity.Front Chem. 2022 Nov 17;10:1008233. doi: 10.3389/fchem.2022.1008233. eCollection 2022. Front Chem. 2022. PMID: 36465862 Free PMC article.
References
-
- Aizenman E., Lipton,S.A. and Loring,R.H. (1989) Selective modulation of NMDA responses by reduction and oxidation. Neuron, 2, 1257–1263. - PubMed
-
- Armstrong N. and Gouaux,E. (2000) Mechanisms for activation and antagonism of an AMPA-sensitive glutamate receptor: crystal structures of the GluR2 ligand binding core. Neuron, 28, 165–181. - PubMed
-
- Armstrong N., Sun,Y., Chen,G.-Q. and Gouaux,E. (1998) Structure of a glutamate receptor ligand binding core in complex with kainate. Nature, 395, 913–917. - PubMed
-
- Brunger A.T. et al. (1998) Crystallography and NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. - PubMed
-
- CCP4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr.D, 50, 760–763. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

