A novel jmjC domain protein modulates heterochromatization in fission yeast
- PMID: 12773576
- PMCID: PMC156127
- DOI: 10.1128/MCB.23.12.4356-4370.2003
A novel jmjC domain protein modulates heterochromatization in fission yeast
Abstract
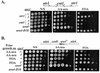
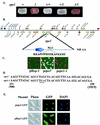
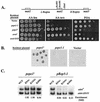
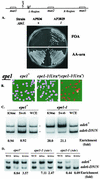
The heterochromatin domain at the mat locus of Schizosaccharomyces pombe is bounded by the IR-L and IR-R barriers. A genetic screen for mutations that promote silencing beyond IR-L revealed a novel gene named epe1, encoding a conserved nuclear protein with a jmjC domain. Disruption of epe1 promotes continuous spreading of heterochromatin-associated histone modifications and Swi6 binding to chromatin across heterochromatic barriers. It also enhances position effect variegation at heterochromatic domains, suppresses mutations in silencing genes, and stabilizes the repressed epigenetic state at the mat locus. However, it does not enhance silencing establishment. Our analysis suggests that the jmjC domain is essential for Epe1 activity and that Epe1 counteracts transcriptional silencing by negatively affecting heterochromatin stability. Consistent with this proposition, the meiotic stability of established heterochromatin beyond IR-L is diminished by Epe1 activity, and overexpression of Epe1 disrupts heterochromatin through acetylation of H3-K9 and H3-K14 and methylation of H3-K4. Furthermore, overexpression of Epe1 elevates the rate of chromosome loss. We propose that Epe1 helps control chromatin organization by down-regulating the stability of epigenetic marks that govern heterochromatization.
Figures





Similar articles
-
Regulation of ectopic heterochromatin-mediated epigenetic diversification by the JmjC family protein Epe1.PLoS Genet. 2019 Jun 17;15(6):e1008129. doi: 10.1371/journal.pgen.1008129. eCollection 2019 Jun. PLoS Genet. 2019. PMID: 31206516 Free PMC article.
-
Swi6/HP1 recruits a JmjC domain protein to facilitate transcription of heterochromatic repeats.Mol Cell. 2006 Jun 9;22(5):681-92. doi: 10.1016/j.molcel.2006.05.010. Mol Cell. 2006. PMID: 16762840
-
An H3K9 methylation-dependent protein interaction regulates the non-enzymatic functions of a putative histone demethylase.Elife. 2020 Mar 20;9:e53155. doi: 10.7554/eLife.53155. Elife. 2020. PMID: 32195666 Free PMC article.
-
Unprogrammed epigenetic variation mediated by stochastic formation of ectopic heterochromatin.Curr Genet. 2020 Apr;66(2):319-325. doi: 10.1007/s00294-019-01031-4. Epub 2019 Oct 9. Curr Genet. 2020. PMID: 31598751 Review.
-
Epigenetic Regulation of Chromatin States in Schizosaccharomyces pombe.Cold Spring Harb Perspect Biol. 2015 Jul 1;7(7):a018770. doi: 10.1101/cshperspect.a018770. Cold Spring Harb Perspect Biol. 2015. PMID: 26134317 Free PMC article. Review.
Cited by
-
Opposing Roles of FACT for Euchromatin and Heterochromatin in Yeast.Biomolecules. 2023 Feb 16;13(2):377. doi: 10.3390/biom13020377. Biomolecules. 2023. PMID: 36830746 Free PMC article. Review.
-
HP1 oligomerization compensates for low-affinity H3K9me recognition and provides a tunable mechanism for heterochromatin-specific localization.Sci Adv. 2022 Jul 8;8(27):eabk0793. doi: 10.1126/sciadv.abk0793. Epub 2022 Jul 8. Sci Adv. 2022. PMID: 35857444 Free PMC article.
-
The euchromatic histone mark H3K36me3 preserves heterochromatin through sequestration of an acetyltransferase complex in fission yeast.Microb Cell. 2020 Jan 3;7(3):80-92. doi: 10.15698/mic2020.03.711. Microb Cell. 2020. PMID: 32161768 Free PMC article.
-
H3K9me-independent gene silencing in fission yeast heterochromatin by Clr5 and histone deacetylases.PLoS Genet. 2011 Jan 6;7(1):e1001268. doi: 10.1371/journal.pgen.1001268. PLoS Genet. 2011. PMID: 21253571 Free PMC article.
-
Functional characterization of Nicotiana benthamiana homologs of peanut water deficit-induced genes by virus-induced gene silencing.Planta. 2007 Feb;225(3):523-39. doi: 10.1007/s00425-006-0367-0. Epub 2006 Aug 22. Planta. 2007. PMID: 16924536
References
-
- Allshire, R. C. 1997. Centromeres, checkpoints and chromatid cohesion. Curr. Opin. Genet. Dev. 7:264-275. - PubMed
-
- Allshire, R. C. 1996. Transcriptional silencing in the fission yeast: a manifestation of higher order chromosome structure and functions, p. 443-466. In V. E. A. Russo, R. A. Martienssen, and A. D. Riggs (ed.), Epigenetic mechanisms of gene regulation. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
-
- Allshire, R. C., J.-P. Javerzat, N. J. Redhead, and G. Cranston. 1994. Position effect variegation at fission yeast centromeres. Cell 76:157-169. - PubMed
-
- Allshire, R. C., E. R. Nimmo, E. Ekwall, J. P. Javerzat, and G. Cranston. 1995. Mutations derepressing silent centromeric domains in fission yeast disrupt chromosome segregation. Genes Dev. 9:218-233. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
