The differential ability of HLA B*5701+ long-term nonprogressors and progressors to restrict human immunodeficiency virus replication is not caused by loss of recognition of autologous viral gag sequences
- PMID: 12768008
- PMCID: PMC156173
- DOI: 10.1128/jvi.77.12.6889-6898.2003
The differential ability of HLA B*5701+ long-term nonprogressors and progressors to restrict human immunodeficiency virus replication is not caused by loss of recognition of autologous viral gag sequences
Abstract
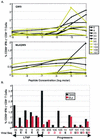
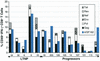
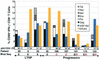
Although the HLA B(*)5701 class I allele is highly overrepresented among human immunodeficiency virus (HIV)-infected long-term nonprogressors (LTNPs), it is also present at the expected frequency (11%) in patients with progressive HIV infection. Whether B57(+) progressors lack restriction of viral replication because of escape from recognition of highly immunodominant B57-restricted gag epitopes by CD8(+) T cells remains unknown. In this report, we investigate the association between restriction of virus replication and recognition of autologous virus sequences in 27 B(*)57(+) patients (10 LTNPs and 17 progressors). Amplification and direct sequencing of single molecules of viral cDNA or proviral DNA revealed low frequencies of genetic variations in these regions of gag. Furthermore, CD8(+) T-cell recognition of autologous viral variants was preserved in most cases. In two patients, responses to autologous viral variants were not demonstrable at one epitope. By using a novel technique to isolate primary CD4(+) T cells expressing autologous viral gene products, it was found that 1 to 13% of CD8(+) T cells were able to respond to these cells by gamma interferon production. In conclusion, escape-conferring mutations occur infrequently within immunodominant B57-restricted gag epitopes and are not the primary mechanism of virus evasion from immune control in B(*)5701(+) HIV-infected patients. Qualitative features of the virus-specific CD8(+) T-cell response not measured by current assays remain the most likely determinants of the differential abilities of HLA B(*)5701(+) LTNPs and progressors to restrict virus replication.
Figures


Similar articles
-
Viral replication capacity as a correlate of HLA B57/B5801-associated nonprogressive HIV-1 infection.J Immunol. 2007 Sep 1;179(5):3133-43. doi: 10.4049/jimmunol.179.5.3133. J Immunol. 2007. PMID: 17709528
-
Correlates of spontaneous viral control among long-term survivors of perinatal HIV-1 infection expressing human leukocyte antigen-B57.AIDS. 2010 Jun 19;24(10):1425-35. doi: 10.1097/QAD.0b013e32833a2b5b. AIDS. 2010. PMID: 20539088 Free PMC article.
-
Efficient processing of the immunodominant, HLA-A*0201-restricted human immunodeficiency virus type 1 cytotoxic T-lymphocyte epitope despite multiple variations in the epitope flanking sequences.J Virol. 1999 Dec;73(12):10191-8. doi: 10.1128/JVI.73.12.10191-10198.1999. J Virol. 1999. PMID: 10559335 Free PMC article.
-
A nonprogressive clinical course in HIV-infected individuals expressing human leukocyte antigen B57/5801 is associated with preserved CD8+ T lymphocyte responsiveness to the HW9 epitope in Nef.J Infect Dis. 2008 Mar 15;197(6):871-9. doi: 10.1086/528695. J Infect Dis. 2008. PMID: 18279072
-
Persistent recognition of autologous virus by high-avidity CD8 T cells in chronic, progressive human immunodeficiency virus type 1 infection.J Virol. 2004 Jan;78(2):630-41. doi: 10.1128/jvi.78.2.630-641.2004. J Virol. 2004. PMID: 14694094 Free PMC article.
Cited by
-
Frequency and phenotype of human immunodeficiency virus envelope-specific B cells from patients with broadly cross-neutralizing antibodies.J Virol. 2009 Jan;83(1):188-99. doi: 10.1128/JVI.01583-08. Epub 2008 Oct 15. J Virol. 2009. PMID: 18922865 Free PMC article.
-
HLA B*5701-positive long-term nonprogressors/elite controllers are not distinguished from progressors by the clonal composition of HIV-specific CD8+ T cells.J Virol. 2012 Apr;86(7):4014-8. doi: 10.1128/JVI.06982-11. Epub 2012 Jan 25. J Virol. 2012. PMID: 22278241 Free PMC article.
-
Control of simian immunodeficiency virus SIVmac239 is not predicted by inheritance of Mamu-B*17-containing haplotypes.J Virol. 2007 Jan;81(1):406-10. doi: 10.1128/JVI.01636-06. Epub 2006 Nov 1. J Virol. 2007. PMID: 17079280 Free PMC article.
-
Effective T-cell responses select human immunodeficiency virus mutants and slow disease progression.J Virol. 2007 Jun;81(12):6742-51. doi: 10.1128/JVI.00022-07. Epub 2007 Apr 4. J Virol. 2007. PMID: 17409157 Free PMC article.
-
HIV-1 evolution following transmission to an HLA-B*5801-positive patient.J Infect Dis. 2009 Dec 15;200(12):1820-4. doi: 10.1086/648377. J Infect Dis. 2009. PMID: 19909081 Free PMC article.
References
-
- Allen, T. M., D. H. O'Connor, P. Jing, J. L. Dzuris, B. R. Mothe, T. U. Vogel, E. Dunphy, M. E. Liebl, C. Emerson, N. Wilson, K. J. Kunstman, X. Wang, D. B. Allison, A. L. Hughes, R. C. Desrosiers, J. D. Altman, S. M. Wolinsky, A. Sette, and D. I. Watkins. 2000. Tat-specific cytotoxic T lymphocytes select for SIV escape variants during resolution of primary viraemia. Nature 407:386-390. - PubMed
-
- Appay, V., D. F. Nixon, S. M. Donahoe, G. M. Gillespie, T. Dong, A. King, G. S. Ogg, H. M. Spiegel, C. Conlon, C. A. Spina, D. V. Havlir, D. D. Richman, A. Waters, P. Easterbrook, A. J. McMichael, and S. L. Rowland-Jones. 2000. HIV-specific CD8+ T cells produce antiviral cytokines but are impaired in cytolytic function. J. Exp. Med. 192:63-76. - PMC - PubMed
-
- Barber, L. D., L. Percival, K. L. Arnett, J. E. Gumperz, L. Chen, and P. Parham. 1997. Polymorphism in the alpha 1 helix of the HLA-B heavy chain can have an overriding influence on peptide-binding specificity. J. Immunol. 158:1660-1669. - PubMed
-
- Barouch, D. H., J. Kunstman, M. J. Kuroda, J. E. Schmitz, S. Santra, F. W. Peyerl, G. R. Krivulka, K. Beaudry, M. A. Lifton, D. A. Gorgone, D. C. Montefiori, M. G. Lewis, S. M. Wolinsky, and N. L. Letvin. 2002. Eventual AIDS vaccine failure in a rhesus monkey by viral escape from cytotoxic T lymphocytes. Nature 415:335-339. - PubMed
-
- Betts, M. R., D. R. Ambrozak, D. C. Douek, S. Bonhoeffer, J. M. Brenchley, J. P. Casazza, R. A. Koup, and L. J. Picker. 2001. Analysis of total human immunodeficiency virus (HIV)-specific CD4+ and CD8+ T-cell responses: relationship to viral load in untreated HIV infection. J. Virol. 75:11983-11991. - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

