Programming peptidomimetic syntheses by translating genetic codes designed de novo
- PMID: 12754376
- PMCID: PMC164450
- DOI: 10.1073/pnas.1132122100
Programming peptidomimetic syntheses by translating genetic codes designed de novo
Abstract
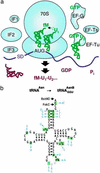
Although the universal genetic code exhibits only minor variations in nature, Francis Crick proposed in 1955 that "the adaptor hypothesis allows one to construct, in theory, codes of bewildering variety." The existing code has been expanded to enable incorporation of a variety of unnatural amino acids at one or two nonadjacent sites within a protein by using nonsense or frameshift suppressor aminoacyl-tRNAs (aa-tRNAs) as adaptors. However, the suppressor strategy is inherently limited by compatibility with only a small subset of codons, by the ways such codons can be combined, and by variation in the efficiency of incorporation. Here, by preventing competing reactions with aa-tRNA synthetases, aa-tRNAs, and release factors during translation and by using nonsuppressor aa-tRNA substrates, we realize a potentially generalizable approach for template-encoded polymer synthesis that unmasks the substantially broader versatility of the core translation apparatus as a catalyst. We show that several adjacent, arbitrarily chosen sense codons can be completely reassigned to various unnatural amino acids according to de novo genetic codes by translating mRNAs into specific peptide analog polymers (peptidomimetics). Unnatural aa-tRNA substrates do not uniformly function as well as natural substrates, revealing important recognition elements for the translation apparatus. Genetic programming of peptidomimetic synthesis should facilitate mechanistic studies of translation and may ultimately enable the directed evolution of small molecules with desirable catalytic or pharmacological properties.
Figures


Similar articles
-
On origin of genetic code and tRNA before translation.Biol Direct. 2011 Feb 22;6:14. doi: 10.1186/1745-6150-6-14. Biol Direct. 2011. PMID: 21342520 Free PMC article.
-
De novo genetic codes and pure translation display.Methods. 2005 Jul;36(3):279-90. doi: 10.1016/j.ymeth.2005.04.011. Methods. 2005. PMID: 16076454
-
Encoding multiple unnatural amino acids via evolution of a quadruplet-decoding ribosome.Nature. 2010 Mar 18;464(7287):441-4. doi: 10.1038/nature08817. Epub 2010 Feb 14. Nature. 2010. PMID: 20154731
-
Synthesis of Peptidyl-tRNA Mimics for Structural Biology Applications.Acc Chem Res. 2023 Oct 3;56(19):2713-2725. doi: 10.1021/acs.accounts.3c00412. Epub 2023 Sep 20. Acc Chem Res. 2023. PMID: 37728742 Free PMC article. Review.
-
Aminoacyl-tRNA synthetases.RNA. 2020 Aug;26(8):910-936. doi: 10.1261/rna.071720.119. Epub 2020 Apr 17. RNA. 2020. PMID: 32303649 Free PMC article. Review.
Cited by
-
In vitro selection of highly modified cyclic peptides that act as tight binding inhibitors.J Am Chem Soc. 2012 Jun 27;134(25):10469-77. doi: 10.1021/ja301017y. Epub 2012 Mar 29. J Am Chem Soc. 2012. PMID: 22428867 Free PMC article.
-
Ribosomal synthesis of dehydroalanine-containing peptides.J Am Chem Soc. 2006 Jun 7;128(22):7150-1. doi: 10.1021/ja060966w. J Am Chem Soc. 2006. PMID: 16734454 Free PMC article.
-
Using the ribosome to synthesize peptidomimetics.F1000 Biol Rep. 2009 Jul 8;1:53. doi: 10.3410/B1-53. F1000 Biol Rep. 2009. PMID: 20948631 Free PMC article.
-
Expanding the amino acid repertoire of ribosomal polypeptide synthesis via the artificial division of codon boxes.Nat Chem. 2016 Apr;8(4):317-25. doi: 10.1038/nchem.2446. Epub 2016 Feb 1. Nat Chem. 2016. PMID: 27001726
-
Multiplexed in vivo His-tagging of enzyme pathways for in vitro single-pot multienzyme catalysis.ACS Synth Biol. 2012 Feb 17;1(2):43-52. doi: 10.1021/sb3000029. ACS Synth Biol. 2012. PMID: 22737598 Free PMC article.
References
-
- Judson, H. F. (1979) The Eighth Day of Creation: Makers of the Revolution in Biology (Simon & Schuster, New York), p. 293.
-
- Fahnestock, S. & Rich, A. (1971) Nature New Biol. 229, 8–10. - PubMed
-
- Fahnestock, S. & Rich, A. (1971) Science 173, 340–343. - PubMed
-
- Noren, C. J., Anthony-Cahill, S. J., Griffith, M. C. & Schultz, P. G. (1989) Science 244, 182–188. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

