Functional distinctions between IMP dehydrogenase genes in providing mycophenolate resistance and guanine prototrophy to yeast
- PMID: 12746440
- PMCID: PMC3367515
- DOI: 10.1074/jbc.M303736200
Functional distinctions between IMP dehydrogenase genes in providing mycophenolate resistance and guanine prototrophy to yeast
Abstract
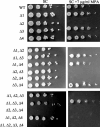
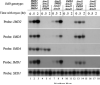
IMP dehydrogenase (IMPDH) catalyzes the rate-limiting step in the de novo synthesis of GTP. Yeast with mutations in the transcription elongation machinery are sensitive to inhibitors of this enzyme such as 6-azauracil and mycophenolic acid, at least partly because of their inability to transcriptionally induce IMPDH. To understand the molecular basis of this drug-sensitive phenotype, we have dissected the expression and function of a four-gene family in yeast called IMD1 through IMD4. We show here that these family members are distinct, despite a high degree of amino acid identity between the proteins they encode. Extrachromosomal copies of IMD1, IMD3, or IMD4 could not rescue the drug-sensitive phenotype of IMD2 deletants. When overexpressed, IMD3 or IMD4 weakly compensated for deletion of IMD2. IMD1 is transcriptionally silent and bears critical amino acid substitutions compared with IMD2 that destroy its function, offering strong evidence that it is a pseudogene. The simultaneous deletion of all four IMD genes was lethal unless growth media were supplemented with guanine. This suggests that there are no other essential functions of the IMPDH homologs aside from IMP dehydrogenase activity. Although neither IMD3 nor IMD4 could confer drug resistance to cells lacking IMD2, either alone was sufficient to confer guanine prototrophy. The special function of IMD2 was provided by its ability to be transcriptionally induced and the probable intrinsic drug resistance of its enzymatic activity.
Figures





Similar articles
-
Dissection of the molecular basis of mycophenolate resistance in Saccharomyces cerevisiae.Yeast. 2005 Nov;22(15):1181-90. doi: 10.1002/yea.1300. Yeast. 2005. PMID: 16278936
-
Regulation of an IMP dehydrogenase gene and its overexpression in drug-sensitive transcription elongation mutants of yeast.J Biol Chem. 2001 Aug 31;276(35):32905-16. doi: 10.1074/jbc.M105075200. Epub 2001 Jul 5. J Biol Chem. 2001. PMID: 11441018 Free PMC article.
-
Detection of the mycophenolate-inhibited form of IMP dehydrogenase in vivo.Proc Natl Acad Sci U S A. 2004 Aug 17;101(33):12171-6. doi: 10.1073/pnas.0403341101. Epub 2004 Aug 3. Proc Natl Acad Sci U S A. 2004. PMID: 15292516 Free PMC article.
-
Consequences of IMP dehydrogenase inhibition, and its relationship to cancer and apoptosis.Curr Med Chem. 1999 Jul;6(7):561-74. Curr Med Chem. 1999. PMID: 10390601 Review.
-
The structure of inosine 5'-monophosphate dehydrogenase and the design of novel inhibitors.Immunopharmacology. 2000 May;47(2-3):163-84. doi: 10.1016/s0162-3109(00)00193-4. Immunopharmacology. 2000. PMID: 10878288 Review.
Cited by
-
Basic mechanisms of RNA polymerase II activity and alteration of gene expression in Saccharomyces cerevisiae.Biochim Biophys Acta. 2013 Jan;1829(1):39-54. doi: 10.1016/j.bbagrm.2012.09.007. Epub 2012 Sep 26. Biochim Biophys Acta. 2013. PMID: 23022618 Free PMC article. Review.
-
Amyloid-like assembly of the low complexity domain of yeast Nab3.Prion. 2015;9(1):34-47. doi: 10.1080/19336896.2014.997618. Epub 2015 Jan 22. Prion. 2015. PMID: 25611193 Free PMC article.
-
Metabolic regulation of IMD2 transcription and an unusual DNA element that generates short transcripts.Mol Cell Biol. 2007 Apr;27(8):2821-9. doi: 10.1128/MCB.02159-06. Epub 2007 Feb 12. Mol Cell Biol. 2007. PMID: 17296737 Free PMC article.
-
Accumulation of unstable promoter-associated transcripts upon loss of the nuclear exosome subunit Rrp6p in Saccharomyces cerevisiae.Proc Natl Acad Sci U S A. 2006 Feb 28;103(9):3262-7. doi: 10.1073/pnas.0507783103. Epub 2006 Feb 16. Proc Natl Acad Sci U S A. 2006. PMID: 16484372 Free PMC article.
-
A novel domain in Set2 mediates RNA polymerase II interaction and couples histone H3 K36 methylation with transcript elongation.Mol Cell Biol. 2005 Apr;25(8):3305-16. doi: 10.1128/MCB.25.8.3305-3316.2005. Mol Cell Biol. 2005. PMID: 15798214 Free PMC article.
References
-
- Jackson RC, Weber G, Morris HP. Nature. 1975;256:331–333. - PubMed
-
- Hedstrom L. Curr. Med. Chem. 1999;6:545–560. - PubMed
-
- Snyder FF, Lightfoot T, Hodges SD. In: Purine and Pyrimidine Metabolism in Man VIII. Sahota A, Taylor M, editors. Plenum Press; New York: 1995. pp. 725–728.
-
- Zhang R, Evans G, Rotella F, Westbrook E, Huberman E, Joachimiak A, Collart FR. Curr. Med. Chem. 1999;6:537–543. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

