Split genes and their expression in Kaposi's sarcoma-associated herpesvirus
- PMID: 12740832
- PMCID: PMC4681429
- DOI: 10.1002/rmv.387
Split genes and their expression in Kaposi's sarcoma-associated herpesvirus
Abstract
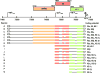
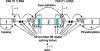
A split or interrupted gene is defined as a gene consisting of introns and exons. Removal (splicing) of the intron(s) from a primary transcript (pre-mRNA) is essential for creating a mRNA. Initial assignment of a potential protein coding region in the KSHV genome was based on the initiation codon context and predicted protein size larger than 100 amino acids, but the gene discontinuity was disregarded. Experimental investigation of the assigned ORFs has demonstrated that there are up to 25 split genes, more than one fourth of the total KSHV genes described in the KSHV genome. This includes the genes involved in all phases (latent, immediate early, early and late) of KSHV infection. The complexity of a split gene expression depends upon the availability of a proximal promoter and polyadenylation (pA) signal. Sharing a single promoter or a single pA signal by two or three genes is not uncommon in the expression of KSHV split genes and the resulting transcripts are usually polycistronic. Among those of KSHV split genes, 15 genes express a bicistronic or tricistronic RNA and 10 genes express a monocistronic RNA. Alternative RNA splicing could happen in a particular pre-mRNA due to intron or exon inclusion or skipping or the presence of an alternative 5' splice site or 3' splice site. This may, respectively, result in at least 8 species of K8 and 14 species of K15 transcripts. This appears to be related to cell differentiation and stages of the virus infection, presumably involving viral cis elements and trans splicing factors.
Figures


Similar articles
-
Kaposi's sarcoma-associated herpesvirus K8beta is derived from a spliced intermediate of K8 pre-mRNA and antagonizes K8alpha (K-bZIP) to induce p21 and p53 and blocks K8alpha-CDK2 interaction.J Virol. 2005 Nov;79(22):14207-21. doi: 10.1128/JVI.79.22.14207-14221.2005. J Virol. 2005. PMID: 16254356 Free PMC article.
-
Kaposi's Sarcoma-Associated Herpesvirus Hijacks RNA Polymerase II To Create a Viral Transcriptional Factory.J Virol. 2017 May 12;91(11):e02491-16. doi: 10.1128/JVI.02491-16. Print 2017 Jun 1. J Virol. 2017. PMID: 28331082 Free PMC article.
-
Identification of Novel Kaposi's Sarcoma-Associated Herpesvirus Orf50 Transcripts: Discovery of New RTA Isoforms with Variable Transactivation Potential.J Virol. 2016 Dec 16;91(1):e01434-16. doi: 10.1128/JVI.01434-16. Print 2017 Jan 1. J Virol. 2016. PMID: 27795414 Free PMC article.
-
Regulation of growth signalling and cell cycle by Kaposi's sarcoma-associated herpesvirus genes.Int J Exp Pathol. 2004 Dec;85(6):305-19. doi: 10.1111/j.0959-9673.2004.00407.x. Int J Exp Pathol. 2004. PMID: 15566428 Free PMC article. Review.
-
[Replication Machinery of Kaposi's Sarcoma-associated Herpesvirus and Drug Discovery Research].Yakugaku Zasshi. 2019;139(1):69-73. doi: 10.1248/yakushi.18-00164-2. Yakugaku Zasshi. 2019. PMID: 30606932 Review. Japanese.
Cited by
-
Protein kinase CK2 phosphorylation regulates the interaction of Kaposi's sarcoma-associated herpesvirus regulatory protein ORF57 with its multifunctional partner hnRNP K.Nucleic Acids Res. 2004 Oct 14;32(18):5553-69. doi: 10.1093/nar/gkh876. Print 2004. Nucleic Acids Res. 2004. Retraction in: Nucleic Acids Res. 2022 Jul 22;50(13):7800. doi: 10.1093/nar/gkac574 PMID: 15486205 Free PMC article. Retracted.
-
Delineation of a core RNA element required for Kaposi's sarcoma-associated herpesvirus ORF57 binding and activity.Virology. 2011 Oct 25;419(2):107-16. doi: 10.1016/j.virol.2011.08.006. Epub 2011 Sep 1. Virology. 2011. PMID: 21889182 Free PMC article.
-
Identification and characterization of the product encoded by ORF69 of Kaposi's sarcoma-associated herpesvirus.J Virol. 2008 May;82(9):4562-72. doi: 10.1128/JVI.02400-07. Epub 2008 Feb 27. J Virol. 2008. PMID: 18305046 Free PMC article.
-
Chromatin immunoprecipitation and microarray analysis suggest functional cooperation between Kaposi's Sarcoma-associated herpesvirus ORF57 and K-bZIP.J Virol. 2013 Apr;87(7):4005-16. doi: 10.1128/JVI.03459-12. Epub 2013 Jan 30. J Virol. 2013. PMID: 23365430 Free PMC article.
-
Genome-Wide Identification and Gene Expression Analysis of the OTU DUB Family in Oryza sativa.Viruses. 2022 Feb 14;14(2):392. doi: 10.3390/v14020392. Viruses. 2022. PMID: 35215984 Free PMC article.
References
-
- Chang Y, Cesarman E, Pessin MS, et al. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi’s sarcoma. Science. 1994;266:1865–1869. - PubMed
-
- Boshoff C, Weiss RA. Kaposi’s sarcoma-associated herpesvirus. Adv Cancer Res. 1998;75:57–86. - PubMed
-
- Cesarman E, Chang Y, Moore PS, et al. Kaposi’s sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N Engl J Med. 1995;332:1186–1191. - PubMed
-
- Soulier J, Grollet L, Oksenhendler E, et al. Kaposi’s sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman’s disease. Blood. 1995;86:1276–1280. - PubMed
-
- Beksac M, Ma M, Akyerli C, et al. Frequent demonstration of human herpesvirus 8 (HHV-8) in bone marrow biopsy samples from Turkish patients with multiple myeloma (MM) Leukemia. 2001;15:1268–1273. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

